-
PDF
- Split View
-
Views
-
Cite
Cite
Anton A. Sanderfoot, Valentina Kovaleva, Haiyan Zheng, Natasha V. Raikhel, The t-SNARE AtVAM3p Resides on the Prevacuolar Compartment in Arabidopsis Root Cells, Plant Physiology, Volume 121, Issue 3, November 1999, Pages 929–938, https://doi.org/10.1104/pp.121.3.929
Close - Share Icon Share
Abstract
Protein cargo is trafficked between the organelles of the endomembrane system inside transport vesicles, a process mediated by integral membrane proteins called SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptors) that reside on the surface of the vesicle (v-SNAREs) and target membrane (t-SNAREs). In examining transport of cargo between the trans-Golgi network and the vacuole in Arabidopsis, we have previously characterized AtPEP12p as a t-SNARE residing on the prevacuolar compartment and AtVTI1a as a v-SNARE that interacts with AtPEP12p. Recently, we have begun to characterize AtVAM3p, another Arabidopsis t-SNARE that shows high sequence homology to AtPEP12p. We have found that AtVTI1a also interacts with AtVAM3p, suggesting a role for this t-SNARE in post-Golgi trafficking. AtVAM3p has been suggested to localize to the vacuolar membrane in Arabidopsis cells; however, using specific antisera and expression of epitope-tagged versions of each t-SNARE, we have discovered that AtVAM3p is found on the same prevacuolar structure as AtPEP12p in Arabidopsis root cells.
Trafficking of cargo between the organelles of the endomembrane system is accomplished through the use of small, membrane-bound transport vesicles. The process by which these vesicles differentiate the correct target membrane from all other organelles is believed to be mediated by integral membrane proteins called SNAREs (solubleN-ethylmaleimide sensitive factor attachment protein receptors) that reside on the surface of the vesicle (v-SNAREs) and target membrane (t-SNAREs; for review, see Sanderfoot and Raikhel, 1999). This process has been best studied in yeast (Saccharomyces cerevisiae) and mammalian cells, although SNAREs are now known in Arabidopsis as well (Bassham et al., 1995; Lukowitz et al., 1996; Sato et al., 1997; Leyman et al., 1999; Zheng et al., 1999a, 1999b).
The prototypical t-SNARE is mammalian syntaxin 1, a protein required for the fusion of synaptic vesicles at the plasma membrane of neural cells (Bennett et al., 1992). In the yeast cell, all of the clearly identifiable t-SNAREs have been characterized (Pelham, 1998). In general, each endomembrane compartment in a yeast cell contains a single syntaxin-type t-SNARE that is responsible for traffic into that compartment, while only the plasma membrane contains more than a single syntaxin-type t-SNARE (for review, see Pelham, 1998). On the other hand, it appears that multiple t-SNAREs reside on the same organelle in eukaryotes such as mammals. Four distinct syntaxin proteins reside on the mammalian plasma membrane (Bennett et al., 1993), while three have been proposed to localize to the late Golgi (Bock et al., 1997; Advani et al., 1998; Tang et al., 1998). Some of these multiple isoforms in mammalian cells appear to represent cell-specific applications, while others are confined to specific domains of a particular compartment or may mediate specific targeting pathways (Low et al., 1996; Galli et al., 1998). Multiple genes encoding homologs of yeast SNAREs are common in plants as well (Zheng et al., 1999a, 1999b). Whether this multiplicity found in plants and mammals represents redundancy, an adaptation to multicellularity, or is the result of the complexity found in mammalian and plant systems remains to be determined.
Some of the components involved in trafficking of vacuolar cargo between the Golgi and prevacuolar compartment (PVC) in Arabidopsis have been characterized (for review, see Miller and Anderson, 1999). Research has indicated that some vacuolar cargo is packaged into clathrin-coated vesicles through the action of the putative vacuolar cargo receptor AtELP/BP-80 (Ahmed et al., 1997; Paris et al., 1997;Sanderfoot et al., 1998). AtELP co-localizes with the v-SNARE AtVTI1a at the trans-Golgi network (Zheng et al., 1999b), and both proteins are probably packaged into clathrin-coated vesicles that travel to the PVC. AtVTI1a is capable of interacting with AtPEP12p, the t-SNARE of the PVC (Zheng et al., 1999b). This interaction probably leads to vesicle fusion and results in the co-localization of all three of these proteins on the PVC (Sanderfoot et al., 1998; Zheng et al., 1999b). Since none of these proteins is found on the vacuolar membrane (Conceição et al., 1997; Sanderfoot et al., 1998; Zheng et al., 1999b), it is probable that AtVTI1a and AtELP are recycled back to the Golgi, while AtPEP12p is retained at the PVC.
A second t-SNARE potentially involved in vacuolar targeting, AtVAM3p, has been characterized in Arabidopsis (Sato et al., 1997). It was suggested that AtVAM3p represented an Arabidopsis homolog of the yeast vacuolar t-SNARE ScVam3p. Consistent with this suggestion, AtVAM3p suppressed the vacuolar morphology and carboxypeptidase Y (CPY) sorting defects of a yeast vam3Δ mutant and was found on the vacuolar membrane in the shoot apical meristem of Arabidopsis (Sato et al., 1997). As first noted by Sato et al. (1997), AtVAM3p is highly homologous to AtPEP12p, and its sequence is more homologous to yeast ScPep12p than to ScVam3p. Consistent with this degree of homology to AtPEP12p and ScPep12p, we have found through biochemical analysis and immunoelectron microscopy that AtVAM3p is a resident of the PVC in Arabidopsis roots.
MATERIALS AND METHODS
Sequence Analysis
Protein sequences for several representative t-SNAREs were acquired from GenBank (AtPEP12p, L41651; AtPLP, U85036; AtSED5p,AF051853; AtSYR1p, AF112864; AtTLG2a, AF067789; AtTLG2b, AF154574; AtVAM3p, U88045; HsSyn1, Q16623; HsSyn5, U26648; HsSyn16, AF038897; ScPep12p, M90395; ScSed5p, X66980; ScSso1p, P32867; ScTlg2p, Z74760; and ScVam3p, U57827). Full-length protein sequences were aligned in the MEGALIGN program (DNASTAR, Madison, WI) using the Jotun Hein algorithm with default parameters (Hein, 1990).
Antibody Production and Purification
Antibodies to the cytosolic domain of AtPEP12p have been described previously (Conceição et al., 1997), and have been found to recognize mainly C-terminal epitopes of AtPEP12p (data not shown). A second polyclonal antiserum raised to amino acids 1 to 129 of AtPEP12p fused to a C-terminal hexa-His tag (AtPEP12[1–129]-H6) was generated as follows: PCR was performed on the cloned cDNA of AtPEP12 (Bassham et al., 1995) to generate EcoRI and NdeI sites at the 5′-end of the open reading frame (ORF) (primer F: 5′-G GAA TTC CAT ATG AGT TTC CAA AGA TCT-3′,EcoRI site underlined, NdeI site in bold; primer R: 5′-GGG TCT TTG TAT GTT TCC ATA GAT TCG C-3′). This product was digested with EcoRI and HindIII (found internal to primer R at the 3′-end of the AtPEP12 ORF), and cloned into similarly digested pBluescript KS (Stratagene, La Jolla, CA) to create pNde-AtPEP12; the sequence of this construct was verified by the Michigan State University Sequencing Facility.
The plasmid pNde-AtPEP12 was digested with NdeI andPvuII (encoding amino acids 1–129 of AtPEP12p), and cloned into the vector pET-23b (Novagen, Madison, WI) prepared by digestion with XhoI, followed by treatment with T4 DNA polymerase (Boehringer Mannheim, Indianapolis) in the presence of dNTPs, heat treatment to inactivate the polymerase, and then digestion withNdeI. Soluble AtPEP12p(1–129)-H6 was overexpressed in Escherichia coli, and protein was purified using a nickel-nitrilotriacetic acid Sepharose column according to the manufacturer's protocol (Novagen). A fusion of amino acids 21 to 200 of AtVAM3p to the C terminus of glutathione-S-transferase (GST) (Sato et al., 1997), was overexpressed in E. coli and purified over a glutathione-Sepharose 4B column according to the manufacturer's protocol (Pharmacia Biotech, Piscataway, NJ). Rabbit polyclonal antibodies were raised to GST-AtVAM3p and AtPEP12(1–129)-H6 by Cocalico Biologicals (Reamstown, PA).
To reduce the cross-reactivity with AtVAM3p found in both types of AtPEP12p-antisera (see “Results”), the antisera raised against AtPEP12p were applied to a glutathione-Sepharose column containing bound GST-AtVAM3p, and the flowthroughs containing antibodies that did not recognize the GST-AtVAM3p were collected. These antibodies retained reactivity with respect to AtPEP12p and had no detectable cross-reactivity to AtVAM3p (see “Results”). AtVAM3p-antiserum was purified in a similar manner by passage over a glutathione-Sepharose column containing GST-AtPEP12p (amino acids 5–257; Zheng et al., 1999b). These antibodies retained reactivity with respect to AtVAM3p and had no detectable cross-reactivity to AtPEP12p (see “Results”).
Yeast Expression of Arabidopsis t-SNARE cDNAs
The cDNAs encoding AtPEP12 (Bassham et al., 1995) orAtVAM3 (Sato et al., 1997) were each cloned into the yeast multicopy plasmid pVT102-U (Vernet et al., 1987). Yeast (Saccharomyces cerevisiae) strain INVSc-1 (Invitrogen, Carlsbad, CA) was transformed with these cDNAs and maintained according to standard yeast protocols (Sherman et al., 1979). Yeast were grown at 27°C in synthetic dextrose medium without uracil (to select for the plasmid) to an OD600 of approximately 1.0. Yeast were lysed by glass bead disruption in yeast lysis buffer (50 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 6.5, 5 mm EDTA, 200 mm sorbitol, and 1 mmdithiothreitol) containing protease inhibitors (100 μm phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 1 μg/mL pepstatin). After clearing the lysate at 2,000g for 10 min at 4°C, the amount of total protein in the supernatant was quantified (Bradford, 1976). Ten micrograms of total protein was separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with antisera to AtPEP12p, AtVAM3p (see above), and yeast CPY (a gift from Dr. Tom Stevens, University of Oregon).
Overexpression of t-SNAREs in Arabidopsis
AtPEP12p containing an N-terminal T7 epitope was constructed as follows: The AtPEP12 cDNA (Bassham et al., 1995) was digested with BglII and HindIII and cloned into pET-23b (Novagen) digested with BamHI andHindIII, resulting in the fusion of the T7 epitope to amino acids 5 to 279 of AtPEP12p. The epitope-tagged cDNA was cloned into the yeast expression vector pVT102-U (Vernet et al., 1987). When expressed in the yeast pep12Δ mutant, T7-AtPEP12 was able to suppress the CPY sorting defect as well as AtPEP12, suggesting that the epitope-tagged protein was functional (data not shown). AtVAM3p was tagged with a T7 epitope as follows: PCR was performed on the cloned cDNA of AtVAM3 (Sato et al., 1997) to generate a BamHI site at the 5′-end of the ORF (primer F: 5′-AGA GGA TTC GCG AAG AAG A-3′, BamHI site in bold; primer R: 5′-CCA GTC ATT GAT GCC TTA-3′). This cDNA was digested with BamHI and EcoRI (found in the vector 3′ to the cDNA) and cloned into similarly digested pET-21a (Novagen), resulting in a fusion of the full-length protein with the T7 epitope. The sequence of T7-AtVAM3 was confirmed by sequencing.T7-AtPEP12, and T7-AtVAM3 were inserted behind the constitutive cauliflower mosaic virus (CaMV) 35S promoter, and transformed into Arabidopsis ecotype Columbia by vacuum infiltration (Bent et al., 1994). Plants were screened by antibiotic resistance, and expression of the epitope-tagged proteins was confirmed using T7-monoclonal antisera (Novagen).
Approximately 2 g of roots of each type of plant were ground separately in 6 mL of lysis buffer (50 mm HEPES, pH 6.5, 5 mm EDTA, 12% [w/v] Suc, and 1 mmdithiothreitol) containing protease inhibitors (see above). The lysate was cleared at 2,000g for 10 min at 4°C, and the supernatant was further centrifuged at 100,000g for 30 min at 4°C. The pellet was resuspended in 0.5 mL of lysis buffer, and protein was quantified according to the method of Bradford (1976). Ten micrograms of microsomal protein was separated by SDS-PAGE, transferred to nitrocellulose, and probed with T7-monoclonal antibodies or the purified AtPEP12p or AtVAM3p antisera.
Co-Immunoprecipitation of T7-AtVTI1a and AtVAM3p
T7-AtVTI1a was immunoprecipitated with a T7-monoclonal antibody from detergent extracts of wild-type or transgenic plants expressingT7-AtVTI1a as previously described (Zheng et al., 1999b). Detergent extracts of wild-type plants or plants expressingT7-AtVTI1a were incubated with T7-monoclonal antibodies cross-linked to agarose beads (Novagen). The flowthrough was collected prior to extensive washing of the beads. Finally, the bound proteins were eluted from the beads with SDS-loading buffer. Equal volumes of the total extract and the flowthrough, together with the equivalent of 100-fold more volume of the eluate, were separated by SDS-PAGE and then probed with either AtPEP12p-specific or AtVAM3p-specific antisera.
Suc Density Gradient Analysis and Electron Microscopy
Microsomes of wild-type (ecotype Columbia) or transgenic Arabidopsis root tissue were prepared as outlined above. The microsome pellet was resuspended in 3.5 mL of lysis buffer with a Dounce homogenizer, then applied to Suc density gradients as described inSanderfoot et al. (1998). Densitometry of the digitized protein blots was performed with imaging software (Image version 1.61, National Institutes of Health, Bethesda, MD). Electron microscopy using cryosections of wild-type or transgenic Arabidopsis roots was essentially as described previously (Sanderfoot et al., 1998; Zheng et al., 1999b).
RESULTS
AtPEP12p and AtVAM3p Show Extensive Sequence Homology
Two t-SNAREs have been hypothesized to be involved in the vacuolar targeting system of Arabidopsis. AtPEP12p was characterized as the product of a cDNA that suppressed the CPY-sorting defect of a yeast mutant that lacks the PVC t-SNARE ScPep12p (Bassham et al., 1995). We have extensively characterized AtPEP12p, both biochemically and through immunogold electron microscopy and found it to reside on the PVC of Arabidopsis root cells (Conceição et al., 1997; Sanderfoot et al., 1998). Similarly, Sato et al. (1997) cloned an Arabidopsis cDNA that suppressed defects found in a yeast mutant that lacks the vacuolar t-SNARE ScVam3p, and referred to the product as AtVAM3p. Through immunogold electron microscopy, Sato et al. (1997) reported that AtVAM3p was found on the tonoplast membrane of the Arabidopsis shoot apical meristem.
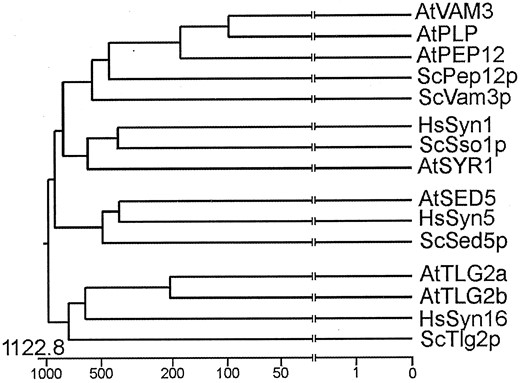
Phylogenetic analysis of selected syntaxin-type t-SNAREs from human (Hs), yeast (Sc), and Arabidopsis (At). Full-length protein sequences were acquired from GenBank (see “Materials and Methods” for accession numbers), aligned, and a phylogenetic tree prepared using the Jotun Hein algorithm of the MEGALIGN program in the DNASTAR package. Below the tree is a scale relating the distance between the sequences. All branches have been truncated (vertical lines) to improve display. See text for details.
On a second branch, two t-SNAREs known to reside on the cis-Golgi and ScSed5p and HsSyn5p (Hardwick and Pelham, 1993; Dascher et al., 1994) cluster with a t-SNARE from Arabidopsis, AtSED5p (A.A. Sanderfoot and N.V. Raikhel, unpublished data). A third cluster consists of HsSyn16p, ScTlg2p, AtTLG2a, and AtTLG2b, t-SNAREs that probably localize to the trans-Golgi network and/or early endosomes (Abeliovich et al., 1998; Holthuis et al., 1998; Séron et al., 1998; Simonsen et al., 1998; A.A. Sanderfoot, V. Kovaleva, and N.V. Raikhel, unpublished observations). Within a fourth cluster, AtPEP12p and AtVAM3p group with a third Arabidopsis t-SNARE, AtPLP (Zheng et al., 1999a). This group of Arabidopsis t-SNAREs as a whole is more homologous to the PVC t-SNARE ScPep12p (Becherer et al., 1996) than to the vacuolar t-SNARE ScVam3p (Wada et al., 1997), and thus lie more closely to ScPep12p on the dendogram. Because AtVAM3p shows a higher degree of homology to a PVC t-SNARE (ScPep12p) than to a vacuolar t-SNARE (ScVam3p), it became important for us to examine its location more closely.
Production of Antisera Specific for AtPEP12p and AtVAM3p
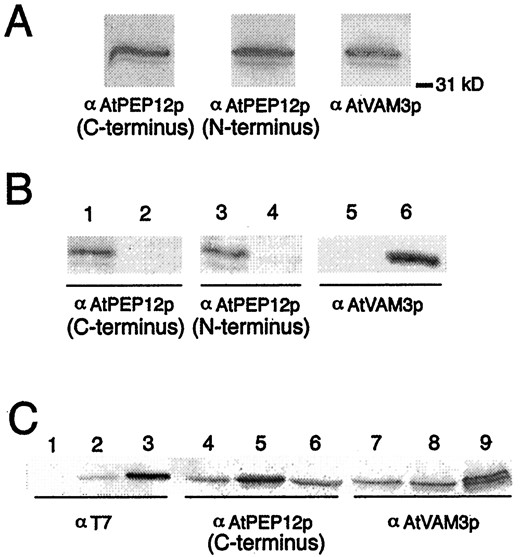
Preparation of antisera specific to AtPEP12p or AtVAM3p. A, Antibodies raised against C- or N-terminal epitopes of AtPEP12p or against the cytoplasmic domain of AtVAM3p recognize bands of similar size (approximately 36 kD) following SDS-PAGE of Arabidopsis microsomal extracts. B, Affinity-purified antisera to each t-SNARE do not cross-react. Extracts from yeast cells expressing the cDNA forAtPEP12 (lanes 1, 3, and 5) or AtVAM3(lanes 2, 4, and 6) were separated by SDS-PAGE, then probed with affinity-purified antisera to either C-terminal-specific AtPEP12p, N-terminal-specific AtPEP12p, or AtVAM3p. C, Expression of epitope-tagged t-SNAREs in transgenic Arabidopsis. Microsomal extracts from wild-type plants (lanes 1, 4, and 7), plants expressingT7-AtPEP12 (lanes 2, 5, and 8), and plants expressingT7-AtVAM3 (lanes 3, 6, and 9) were separated by SDS-PAGE, and then probed with T7 monoclonal antibodies or with affinity-purified antisera to either C-terminal-specific AtPEP12p or AtVAM3p.
A second antiserum generated to epitopes in the N terminus of AtPEP12p (see “Materials and Methods”) showed a high degree of cross-reactivity with AtVAM3p (data not shown). To remove this cross-reactivity, the AtPEP12p antisera were depleted of AtVAM3p epitopes by absorption with GST-AtVAM3p (see “Materials and Methods”). Both of the purified AtPEP12p antisera reacted strongly with the extracts from AtPEP12-expressing yeast cells, but showed no reactivity to extracts from AtVAM3-expressing cells (Fig. 2B, left and center). The AtVAM3p antiserum reacted strongly with protein extracted from AtVAM3-expressing cells, showing very low levels of cross-reactivity withAtPEP12-extracts (data not shown). This low level of cross-reactivity was removed by adsorption with GST-AtPEP12p (Fig. 2B), resulting in specific antiserum for AtVAM3p. No bands were seen for any antisera with yeast extracts that did not express any t-SNARE from Arabidopsis, indicating that none of the antisera cross-reacts with the yeast t-SNARE homologs (data not shown). As a control for equal loading, these same blots were probed with antibodies to the yeast protein CPY (data not shown). These results suggested that the purified antisera for each of these t-SNAREs was specific to their respective antigens.
To ensure that we could unambiguously differentiate AtPEP12p and AtVAM3p, we separately epitope tagged both AtPEP12p and AtVAM3p with a T7-epitope tag (Novagen) and individually expressed the cDNAs under the control of the constitutive CaMV 35S promoter in Arabidopsis plants. It is unlikely that the epitope tag affected the function or localization of either of these proteins, because expression ofT7-AtPEP12 in yeast pep12Δ mutants suppressed the CPY sorting defect to the same extent as untaggedAtPEP12 (data not shown), and the intracellular localization of both epitope-tagged proteins was identical to the endogenous proteins (Fig. 4).
Extracts of transgenic plants expressing either T7-AtPEP12or T7-AtVAM3 probed with T7-monoclonal antibodies showed a single band of approximately 36 or approximately 37 kD, respectively (Fig. 2C, left). In each case, the band recognized by the T7-monoclonal antibodies was also recognized by either the AtPEP12p or AtVAM3p antisera (Figs. 2C, center and right, respectively). Also, the AtPEP12p-specific antiserum did not recognize T7-AtVAM3p, nor did AtVAM3p-specific antiserum recognize T7-AtPEP12p (Fig. 2C, compare lanes 5 and 8, and lanes 6 and 9), again indicating that the purified antisera are specific for each t-SNARE. In each case, the T7-tagged protein was overexpressed with respect to the endogenous proteins (see center and right panels of Fig. 2C); in these particular lines, T7-AtVAM3p was expressed at a higher level than T7-AtPEP12p. We observed some heritable root phenotypes such as reduced growth and excessive branching in some lines overexpressing these T7-tagged t-SNAREs (data not shown); however, the transgenic lines shown were indistinguishable from wild-type plants with respect to root growth and development.
AtVAM3p and AtVTI1a Interact in Vivo
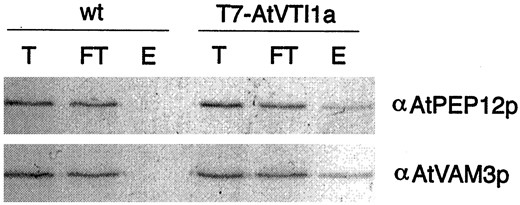
In the yeast cell, the v-SNARE ScVti1p is an important regulator of many distinct pathways to the yeast vacuole (Fischer von Mollard and Stevens, 1999). Not only does ScVti1p interact with ScPep12p as part of trafficking cargo such as CPY to the PVC (Fischer von Mollard and Stevens, 1997), but it also interacts with ScVam3p on the vacuole as part of delivery of two other classes of vacuolar cargo (alkaline phosphatase [ALP] and aminopeptidase I) in distinct trafficking pathways (Fischer von Mollard and Stevens, 1999). Homologs of ScVti1p have been found in Arabidopsis, one of which (AtVTI1a) has been shown to interact in vivo with AtPEP12p (Zheng et al., 1999b). Since ScVti1p interacts with both ScPep12p and ScVam3p, we attempted to address whether AtVTI1a would interact with AtVAM3p. Furthermore, previous experiments investigating the AtVTI1a-AtPEP12p interaction used the unpurified AtPEP12p antiserum, which was found to cross-react with AtVAM3p (see above). For these reasons, we examined the interaction of AtVTI1a with AtPEP12p or AtVAM3p using our specific antisera to each t-SNARE.
AtVAM3p and AtVTI1a interact in vivo. A detergent extract of wild-type (wt) or transgenic Arabidopsis plants expressingT7-AtVTI1a was immunoprecipitated with T7-monoclonal antibodies. Shown are aliquots of the total extract (T), the flowthrough (FT), and the elution (E) from the immunoprecipitation separated by SDS-PAGE, then probed with AtPEP12p-specific or AtVAM3p-specific antiserum.
AtVAM3p Co-Fractionates with AtPEP12p during Density Gradient Analysis of Arabidopsis Root Microsomes
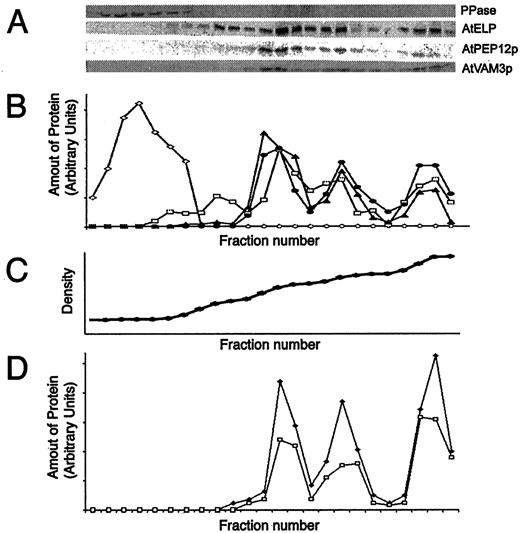
AtVAM3p co-fractionates with PVC markers in Suc density gradients. A, Microsomal extracts of wild-type Arabidopsis plants were separated on Suc density gradients. Twenty-four fractions were taken, TCA precipitated, resuspended in SDS sample buffer, and equal volumes separated by SDS-PAGE. Strips of these blots were probed with antisera specific to: H+-pyrophosphatase (PPase), AtELP, C-terminal specific AtPEP12p, or AtVAM3p. B, These blots were digitized on a flat-bed scanner, and densitometry was performed with imaging software. Shown is a quantification of each fraction (relative to the total amount of each protein loaded onto the gradient) for H+-pyrophosphatase (PPase, ⋄), AtELP (■), AtPEP12p (▴), and AtVAM3p (●). C, The density profile of this gradient was determined by refractometry and is virtually linear. D, Microsomal extracts of plants either expressing T7-AtPEP12 (■) orT7-AtVAM3 (▴) were separated on Suc density gradients as described for A, and quantified as described in B. The density profile of each gradient was similar to that shown in C.
Considering that AtPEP12p was never detected in fractions enriched in tonoplast membranes (Fig. 4A; Sanderfoot et al., 1998) or in purified tonoplast vesicles (Conceição et al., 1997), and that these initial antibodies cross-reacted with AtVAM3p, it seemed that either the in vivo level of cross-reactivity with AtVAM3p was too low to detect AtVAM3p on the vacuole or that AtVAM3p was present in the same membrane fractions as AtPEP12p. To address this question, we examined the fractionation pattern of AtVAM3p using the purified AtVAM3p antiserum and found that AtVAM3p fractionated with three major peaks of 1.12, 1.14, and 1.17 g mL−1 (Fig. 4A), each of which co-fractionated with AtPEP12p. No AtVAM3p was detected in low-density fractions containing the tonoplast marker H+-pyrophosphatase (Maeshima and Yoshida, 1989), indicating that AtVAM3p does not reside on the tonoplast of Arabidopsis roots, and instead appears to reside on the PVC. Similar fractionation patterns were found upon fractionation of green tissues of Arabidopsis, suggesting that this localization is not confined to root tissue (data not shown).
As confirmation of the above results, we again used the transgenic plants expressing T7-AtPEP12 or T7-AtVAM3 to clearly distinguish each t-SNARE. Extracts of these transgenic roots were subjected to Suc density gradient analysis and the fractionation pattern of the T7-tagged t-SNAREs were compared with that of the endogenous proteins. As seen in Figure 4D, T7-AtPEP12p has a fractionation pattern similar to that seen previously for AtPEP12p (see Fig. 4A; Sanderfoot et al., 1998), confirming that our previous results reported using antibodies raised against AtPEP12p (Conceição et al., 1997; Bassham and Raikhel, 1998;Sanderfoot et al., 1998; Zheng et al., 1999b) were not simply due to cross-reactivity with AtVAM3p. Figure 4D shows that T7-AtVAM3p also had a fractionation pattern identical to that seen for AtPEP12p (and to that seen for endogenous AtVAM3p), strongly suggesting a prevacuolar localization for this t-SNARE in root tissue.
AtVAM3p Is Localized on the PVC of Arabidopsis Root Cells
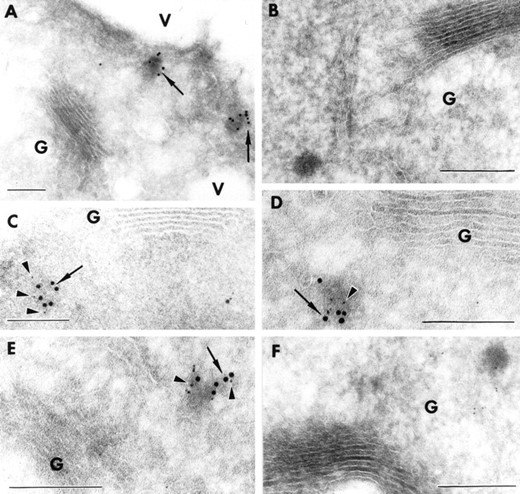
AtVAM3p localizes to the PVC in Arabidopsis root cells. Cryosections of the root tip of transgenic Arabidopsis seedlings expressing either T7-AtPEP12 or T7-AtVAM3were prepared and stained with T7-monoclonal or specific antisera to AtPEP12p (C-terminal specific) or AtVAM3p. In all cases, AtVAM3p is represented by 10-nm gold particles (arrows), while AtPEP12p is represented by 5-nm gold particles (arrowheads). A, Cryosections ofT7-AtVAM3-expressing transgenic plants stained with T7-monoclonal antibodies detected with 10-nm gold. B, Cryosections ofT7-AtVAM3-expressing transgenic plants stained with no antisera followed by 10-nm gold. C, Cryosections ofT7-AtVAM3-expressing transgenic plants stained with T7-monoclonal antibodies detected with 10-nm gold and with affinity-purified AtPEP12p antiserum (C-terminal specific) detected with 5-nm gold. D and E, Cryosections ofT7-AtPEP12-expressing transgenic plants stained with T7-monoclonal antibodies detected with 5-nm gold and with affinity-purified AtVAM3p antiserum detected with 10-nm gold. F, Cryosections of T7-AtPEP12-expressing transgenic plants stained with no antibodies followed by 10-nm gold and preimmune serum for AtPEP12p detected with 5-nm gold. Bar = 500 nm.
To determine whether this AtVAM3p-labeled, electron-dense structure was the same PVC as previously described for AtPEP12p, we examined cryosections of transgenic Arabidopsis roots expressing eitherT7-AtVAM3 or T7-AtPEP12 by double-immunolabeling. In these experiments, we found extensive co-localization of AtPEP12p and AtVAM3p on the same PVC (Fig. 5, C–E). Figure 5C shows cryosections of plants expressing T7-AtVAM3 stained with T7-monoclonal antibodies detected with 10-nm gold particles, followed by AtPEP12p-specific antisera detected with 5-nm gold particles. Similarly, cryosections of transgenic plants expressingT7-AtPEP12 were stained with T7-monoclonal antibodies detected with 5-nm gold particles, followed by AtVAM3p-specific antisera detected with 10-nm gold particles (Fig. 5, D and E). In cryosections of wild-type plants, we also saw co-localization on PVC-like structures when affinity-purified AtPEP12p antiserum detected with 5-nm gold and AtVAM3p antiserum detected with 10-nm gold were used (data not shown). When T7-monoclonal antibodies were omitted, or if preimmune serum for AtPEP12p or AtVAM3p were used in these experiments, no staining of PVC-like structures was observed (Fig. 5F). Based on these results, we believe that AtVAM3p and AtPEP12p reside on the same PVC in Arabidopsis root cells.
DISCUSSION
According to the yeast model (Pelham, 1998), each of the organelles of the endomembrane system contains a single syntaxin-type t-SNARE that functions to receive traffic into that compartment. This model is based upon the fact that the entire genome sequence has only revealed eight ORFs that are recognizable as syntaxin-type t-SNAREs, each of which has been localized to a different endomembrane compartment (Holthuis et al., 1998; Pelham, 1998). Based upon expressed sequence tag data alone, 16 syntaxin-type t-SNAREs have been defined in mammalian cells (Bock and Scheller, 1997). Similarly, 10 are known in Arabidopsis either as cloned cDNAs or as hypothetical ORFs in the currently available genomic sequence (Bassham et al., 1995; Lukowitz et al., 1996; Sato et al., 1997; Leyman et al., 1999; Zheng et al., 1999a; A.A. Sanderfoot and N.U. Raikhel, unpublished observations).
These observations suggest that multicellular eukaryotes have many more t-SNAREs than yeast. While it is possible that plants and animals may contain additional endomembrane compartments, this seems unlikely. Rather, it appears that plants and animals contain more than a single t-SNARE on each compartment. Whether this multiplicity is due to redundancy, cell-specific expression, or multiple parallel pathways is currently being investigated by several researchers. In the present study, we have presented evidence that the PVC of Arabidopsis roots contains two distinct t-SNAREs, AtPEP12p and AtVAM3p.
In the yeast cell, the vacuolar system contains two t-SNAREs that are required for the efficient targeting of proteins to the vacuole: ScPep12p on the PVC and ScVam3p on the vacuole (Becherer et al., 1996;Wada et al., 1997). Three main routes are taken by different classes of vacuolar cargo, each in a distinct type of transport vesicle (for review, see Bryant and Stevens, 1998). The first class of cargo, typified by CPY, contains N-terminal sorting signals that are recognized by cargo receptors (i.e.: ScVps10p) in the lumen of the trans-Golgi network (Marcusson et al., 1994). This class of cargo travels first to the PVC in a process mediated by ScPep12p (Becherer et al., 1996), and then in a second step to the vacuole in a process mediated by ScVam3p (Wada et al., 1997).
A second class of cargo, typified by ALP, bypasses the PVC and is transported directly to the vacuole in a process also mediated by ScVam3p (Darsow et al., 1997; Piper et al., 1997). Cargo in this class are generally membrane proteins that contain specific peptide motifs (called acidic di-Leu motifs) in their cytoplasmic domains, which lead to the packaging of this class of cargo into vesicles distinct from those of the CPY pathway (Vowels and Payne, 1998). Interestingly, ScVam3p itself contains these sorting motifs and is targeted to the vacuole through the ALP pathway (Darsow et al., 1998). A third pathway involves proteins such as aminopeptidase I, which are synthesized on cytoplasmic ribosomes and transported into the vacuole directly from the cytoplasm in constitutive autophagic vesicles; this process is also mediated by ScVam3p (Darsow et al., 1997). ScVam3p also has a role in vacuole assembly, as it has also been found to be responsible for homotypic fusion of vacuolar membranes (Nichols et al., 1997). Thus, as many as four distinct processes in yeast require the activity of ScVam3p.
Interestingly, the role of AtVAM3p has been examined for only two of these ScVam3p-mediated processes, CPY transport and vacuolar assembly (Sato et al., 1997). Expression of AtVAM3 in yeastvam3Δ mutants suppresses the CPY-sorting and vacuolar morphology defects of this mutant (Sato et al., 1997). Examination of the peptide sequence of AtVAM3p does not reveal any sequence resembling the cytoplasmic sorting motif used to target proteins (including ScVam3p) through the ALP-pathway to the vacuole. Thus, it seems unlikely that AtVAM3p can be targeted to the yeast vacuole in the same manner as ScVam3p. For this reason, it is surprising that AtVAM3p can functionally replace ScVam3p when expressed in vam3Δ mutants. Expression of AtPEP12 in the vam3Δ mutant does not suppress any of the defects found in these mutants (Sato et al., 1997), nor does expression of AtVAM3 in yeastpep12Δ mutants restore the CPY-sorting defects found in these mutant cells (Sato et al., 1997; A.A. Sanderfoot and N.U. Raikhel, unpublished results). Thus, although both of these Arabidopsis t-SNAREs share greater than 65% protein identity, in yeast they are found to play different roles in the vacuolar sorting system. Whether this functional distinction will also be found in plant cells remains to be elucidated.
Do AtPEP12p and AtVAM3p have the same or distinct functions? The yeast complementation results suggest that the role of AtPEP12p is similar to ScPep12p in receiving trans-Golgi network-derived vesicles at the PVC, while AtVAM3p may have a role in vacuolar assembly (as suggested bySato et al., 1997), perhaps in the fusion of multiple PVC compartments into the large central vacuole. By examining the shoot apical meristem of Arabidopsis plants, Sato et al. (1997) reported that AtVAM3p was found only at the junction of two vacuolar membranes, and proposed that this suggests a role in vacuolar assembly. On the other hand, we did not find AtVAM3p on the vacuolar membrane in Arabidopsis root cells. Instead, we found that AtVAM3p is exclusively a resident of the PVC. These results may be relevant considering the distinct cell types examined in these two studies. The cells of the shoot apical meristem contain many small vacuoles, while those of the root cells we examined generally contain only a single, large central vacuole. Thus, it may be that AtVAM3p performs distinct roles in different cell types, or that, developmentally, the fusion of small vacuoles in the shoot apical meristem is equivalent to the fusion of PVCs to each other or to the vacuole in mature vegetative cells. Clearly, more study is required to classify the nature of the PVC and vacuolar compartments of plant cells.
In these studies, we have used epitope-tagged proteins expressed in transgenic plants to clearly distinguish between two highly homologous proteins. While it is possible that either overexpression or the presence of an epitope tag can affect the localization or the function of a protein, we do not feel that this is the case in our studies. First of all, we attempted to avoid problems related to protein overexpression by choosing lines with low to moderate levels of expression of the epitope-tagged proteins (see Fig. 1). Second, we addressed the function of the epitope-tagged proteins using expression in yeast mutants. We found that expression of T7-AtPEP12 in the yeast pep12Δ mutant was able to restore proper sorting of CPY, suggesting that the presence of the epitope tag does not affect the function of AtPEP12p. Third, we examined the intracellular localization of the epitope-tagged proteins with respect to the endogenous proteins. Both by biochemical analysis in Suc density gradients and immunogold electron microscopy, we found that T7-AtPEP12p and T7-AtVAM3p have a staining pattern identical to that of the endogenous (untagged) proteins (see Figs. 4 and 5). We also believe that these epitope-tagged proteins retain their function with respect to protein-to-protein interactions. For example, immunoprecipitation of endogenous AtPEP12p with specific antibodies or of T7-AtPEP12p with T7-monoclonal antibodies, both resulted in the co-immunoprecipitation of AtVTI1 (A.A. Sanderfoot and N.V. Raikhel, unpublished observations). Thus, we feel that these particular epitope-tagged proteins (T7-AtPEP12p and T7-AtVAM3p) reflect the localization and function of the endogenous proteins.
In conclusion, we have found that AtVAM3p is a resident of the PVC in Arabidopsis root cells. Considering these results, is it possible to explain the results reported earlier on the expression of these Arabidopsis t-SNAREs in various yeast mutants? Perhaps in the yeastvam3Δ mutants, AtVAM3p functions in a transport step between the PVC and vacuole, where it replaces one of the functions of ScVam3p, allowing suppression of the defects of this mutant. If this is the case, then AtVAM3p is not functionally equivalent to ScVam3p in all of the functions discovered for this t-SNARE in yeast. Based on protein sequence and intracellular localization, AtVAM3p does not appear to be a true homolog of ScVam3p, and therefore it may be useful to rename it t-SNARE, reserving “VAM3” for the as-yet-unidentified vacuolar t-SNARE in Arabidopsis. However, there is likely to be a limit on how much can be learned from expression of these t-SNAREs in heterologous systems. More research into the role of AtVAM3p in plants is required before we can clearly delineate the precise function of the Pep12p-like t-SNAREs from Arabidopsis.
ACKNOWLEDGMENTS
The authors thank Masa Sato and Yoh Wada for providing the cloned cDNA of AtVAM3 and the plasmid encodingGST-AtVAM3, Tom Stevens for antiserum to CPY, and Weiqing Zeng for assistance in preparing T7-epitope-tagged AtVAM3. We also acknowledge Diane Bassham and John Froehlich for helpful comments on the manuscript.
LITERATURE CITED
Author notes
A.A.S. is a National Institutes of Health Postdoctoral Fellow (no. GM18861). N.V.R. is supported by research grants from the National Science Foundation (grant no. MCB–9507030) and the Department of Energy (grant no. DE–FG02–91ER–20021).
Corresponding author; e-mail nraikhel@pilot.msu.edu; fax 517–353–9168.