-
PDF
- Split View
-
Views
-
Cite
Cite
Sophie Le Pogam, Amritha Seshaadri, Alan Kosaka, Sophie Chiu, Hyunsoon Kang, Steven Hu, Sonal Rajyaguru, Julian Symons, Nick Cammack, Isabel Nájera, Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients, Journal of Antimicrobial Chemotherapy, Volume 61, Issue 6, June 2008, Pages 1205–1216, https://doi.org/10.1093/jac/dkn085
Close - Share Icon Share
Abstract
To characterize the effect of hepatitis C virus (HCV) polymerase intrinsic genetic heterogeneity on the inhibitory activity of nucleoside and non-nucleoside HCV polymerase inhibitors.
The sensitivity of genotype (GT) 1 HCV NS5B clinical isolates from treatment-naive patients to nucleoside and non-nucleoside polymerase inhibitors was assessed. The genetic diversity at the population level, as well as that of their quasispecies, was correlated with the observed reduced sensitivity to inhibitors.
R1479 and NM107 (nucleoside analogues that have entered Phase 2 clinical trials as prodrugs R1626 and NM283, respectively) were similarly active across the tested clinical isolates. Resistance mutations to nucleoside analogues were not observed in any of the isolates. However, the activity of the non-nucleoside thumb II inhibitor NNI-1, palm I inhibitors NNI-2 and NNI-3, and palm II inhibitor HCV-796 was reduced across different isolates. This reduction in inhibitory activity for non-nucleoside inhibitors (NNIs) was, in most cases, correlated with the existence of known NNI resistance mutations in the NS5B polymerase population of the clinical isolates, as detected by population sequencing. Resistance mutations to NNIs were also observed at a low frequency within the clinical isolates' viral quasispecies that allowed for their rapid selection upon drug selective pressure.
The higher frequency of known NNI resistance mutations or polymorphisms known to affect their antiviral potency when compared with the lack of detection of resistance mutations to the nucleoside analogues suggests a potential for primary reduced responsiveness as well as faster development of clinically significant resistance.
Introduction
Hepatitis C virus (HCV), a positive-strand RNA virus, is a member of the genus Hepacivirus in the Flaviviridae family and is the leading cause of liver disease worldwide. It is estimated that over 170 million individuals are infected with HCV.1 The current standard of care provides good clinical efficacy in patients infected with genotypes (GTs) 2 and 3, but is less efficacious in the most prevalent GT 1-infected patients, thereby emphasizing the urgent need for more effective specific targeted antiviral therapies for HCV.2,3
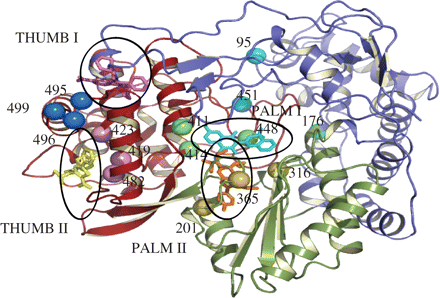
The HCV RNA-dependent RNA polymerase (RdRp) is an essential enzyme for viral RNA replication and therefore an attractive therapeutic target. This enzyme has a crystal structure with the shape of an encircled right hand, and in addition to the active site, four allosteric sites have been identified: palm I (palm domain near the active site), palm II (partially overlapping palm I and towards the active site), thumb I (thumb domain near the fingertips) and thumb II (the outer surface of the thumb domain) (Figure 1). There are a number of polymerase inhibitors that have been advanced to Phase 2 clinical trials and have demonstrated clinical efficacy in monotherapy: nucleoside analogues NM283 (1.2 log drop in viral load at 800 mg twice daily)4 and R1626 (3.7 log drop in viral load at 4500 mg twice daily)5 and the non-nucleoside inhibitor (NNI) HCV-796 that binds to the palm II domain of the polymerase (1.4 log drop in viral load at ≥500 mg twice daily at day 4 followed by viral load rebound while on treatment). Other NNIs that inhibit the isolated enzyme and HCV replication in the replicon system include benzothiadiazines6,7 and acylpyrrolidines8,9 that bind to the palm I site, benzimidazoles and indoles that bind to the thumb I site,10–12 and thiophene carboxylic acids that bind to the thumb II site.13,14 The emergence of resistance has been observed in clinical trials for the protease inhibitor VX-95015 and for the polymerase inhibitor HCV-79616,17 and can be a limiting factor for the efficacy of antiviral therapy. Here, we describe the development of an HCV NS5B phenotypic assay that allows for the monitoring of resistance during clinical trials, assessing the drug sensitivity of the NS5B viral population as observed in the clinical isolates. The effect of HCV genetic diversity on the inhibitory potency of NS5B polymerase inhibitors was investigated by studying GT 1 clinical isolates from treatment-naive patients. This study shows that nucleoside analogues R1479 and NM107 (parent compounds of R1626 and NM283, respectively) were active across genetically diverse clinical isolates. However, the activity of the non-nucleoside thumb II inhibitor NNI-1, palm I inhibitors NNI-2 and NNI-3, and palm II inhibitor HCV-796 was reduced for some isolates, which correlated with the presence of NNI resistance mutations within the viral population. Clonal sequence analysis of the NS5B coding region was performed among clinical isolates and demonstrated the existence of NNI resistance mutations within the quasispecies of untreated patients that could facilitate the rapid selection of drug-resistant variants upon drug pressure.
Overall structure of HCV NS5B polymerase. The structure of NS5B highlights the fingers domain (blue), palm domain (green) and thumb domain (red). Inhibitor-binding positions for thumb I (pink, benzimidazole), thumb II (yellow, NNI-1), palm I (cyan, NNI-2) and palm II (orange, HCV-796) inhibitors are circled. The locations of primary resistance mutations are indicated by spheres for thumb I (blue), thumb II (pink), palm I (green) and palm II (gold) sites. Of the palm I site resistance mutations, those not in direct contact with the inhibitor are coloured cyan.
Materials and methods
Clinical isolates
Ninety-two HCV NS5B clinical isolates were obtained from serum samples of untreated GT 1 HCV-infected individuals. Forty-five samples were from the Phase Ib R1626 study and originated from Australia.5 Others were obtained from New Zealand (39) and from a variety of commercial sources (7 from the USA and 1 from Germany).
Plasmid construction
The Con1-adapted transient replicon (rep PI-luc/ET) and cured Huh-7 cells were obtained from Ralf Bartenschlager (Department of Molecular Virology, University of Heidelberg, Germany).18 The transient replicon repPI-luc/ET vector was modified to replace the pBR322 backbone with the pUC18 backbone and to include two restriction sites flanking the start (AsiSI) and the end (RsrII or SacII) of a truncated NS5B gene. Briefly, the transient replicon includes the poliovirus internal ribosome entry site (IRES), which controls the translation of the firefly luciferase gene. Downstream of the firefly luciferase gene, the IRES from the encephalomyocarditis virus controls the translation of the HCV non-structural genes (NS3, NS4A, NS4B, NS5A and a truncated NS5B). Only vectors containing a full-length NS5B clinical isolate are able to replicate.
The transient replicon containing the GT 1a H77 sequence was adapted from the GT 1b transient replicon by replacing the non-structural region of the Con1 by the H77 sequence, except the first 75 amino acids of NS3 that remained of Con1 origin. For better replication efficiency, three adaptive mutations were introduced.19 Restriction sites AsiSI and RsrII were also introduced at the 5′ and 3′ end of the NS5B gene. All constructs were confirmed by double-stranded DNA sequencing.
HCV RNA extraction and NS5B amplification
HCV RNA was extracted from 400 µL of serum from HCV-infected patients using the ZR Whole-Blood Total RNA kit following manufacturer's instructions (Zymo Research, Orange, CA, USA; catalogue no. R1020). Reverse transcription (RT) was carried out with 5 µL of total RNA and the Taqman RT reagents kit following manufacturer's instructions (ABI, Foster City, CA, USA; catalogue no. N808-0234) using a 29-mer oligo (dA) as primer for RT initiation. NS5B amplification was carried out using GC-RICH PCR system (Roche Diagnostics, Indianapolis, IN, USA; catalogue no. 12 140 306 001) as follows: 4 µL of cDNA was used in a 50 µL reaction containing 28 µL of water, 5 µL of Buffer #3 (1× final), 10 µL of Buffer #2 (1× final), 0.5 µL of forward and reverse primers (0.2 µM final each), 1 µL of dNTPs (0.02 mM final) and 1 µL of enzyme (2 U). PCR reactions were performed as follows: 2 min at 94°C, then 40 cycles of 30 s at 92°C, 1 min at 50°C and 2 min at 72°C, followed by a 5 min extension at 72°C. A nested amplification was carried out in duplicate using the same conditions with primers specific for GT 1a or GT 1b sequences that contained the restriction site sequences for AsiSI and RsrII (or SacII). After restriction enzyme digestion, GT 1b-amplified products were cloned into the GT 1b shuttle replicon vector, whereas GT 1a-amplified products were cloned into the GT 1a shuttle replicon vector. After overnight ligation and transformation, 96 individual colonies were pooled for each clinical isolate. In vitro-transcribed RNA was then prepared using RiboMAX™ T7 Express (Promega, Madison, WI, USA; catalogue no. R1320).
To determine the error rate (ER) of the PCR amplification procedure, the NS5B region of the H77 DNA plasmid was amplified in two independent experiments following the same protocol as the one used for the amplification of clinical isolates. After TA cloning (TOPO TA cloning vector Invitrogen, Carlsbad, CA, USA; catalogue no. 45-0641), two sets of 29 and 28 clones, respectively, were analysed by sequencing. In each set, 10 nucleotide substitutions were found. The ER was determined using the following formula: ER = number of mutation per base pair/number of PCR cycles. For both sets, the ER was calculated at 2.4 × 10−6/bp/PCR cycle.
Sequencing of NS5B polymerase genes
Sequencing spanning the entire NS5B coding region was performed using primers covering both DNA strands, both populations (direct PCR sequencing) or molecular clones sequencing. Sequencing reactions were performed using ABI 3730 xl DNA Analyzer and chromatograms were analysed using Sequencher and VNTI software.
Compounds
Compounds R1479 (4′azido-cytidine), NM107 (2′-C-Me-C), thiophene-2-carboxylic acid (NNI-1), benzothiadiazine (NNI-2) and HCV-796 (benzofuran carboxamide) were synthesized at Roche Palo Alto LLC. Thiazine (NNI-3) was synthesized at Array BioPharma. Stocks of 10 mM concentration were prepared in 100% DMSO and stored at −20°C.
IC50 determinations
For IC50 determination, 4 million cured Huh7 cells were transfected with 5 µg (or 10 µg) of in vitro-transcribed replicon RNA from GT 1b or GT 1a reference strains or from clinical isolate-derived transient replicons. After electroporation, cells were resuspended in 12 mL (or 8 mL) of Dulbecco's modified Eagle's medium (DMEM) containing 5% (v/v) fetal bovine serum and plated in 96-well plate at 30 000 cells/well (or in 48-well plate at 100 000 cells/well, in 200 µL final volume). Compounds (or medium as a control) were added 24 h post-transfection in 3-fold dilutions at a final DMSO concentration of 1% (v/v). Firefly luciferase reporter signal was read 72 h after addition of compounds using the Luciferase Assay system (Promega; catalogue no. E1501). The IC50 values were assessed as the compound concentration at which a 50% reduction in the level of firefly luciferase reporter was observed when compared with control samples in the absence of compound. Dose–response curves and IC50 values were generated by using the XLfit3 program (ID Business Solutions Ltd, Surrey, UK). The 95% CI was calculated on the basis of 2 × SEM. The variability of the assay was represented as the SD as well as the maximum fold changes relative to the mean IC50.
For Con1 and H77 wild-type replicons, the signal-to-noise ratio was 1200- and 2000-fold above background, respectively. Signal-to-noise ratio for the majority of the GT 1b clinical isolates was between 50- and 180-fold and between 50- and 2700-fold for the GT 1a clinical isolates.
Amplification and sequencing of the coding region from replicating transient replicons
Transfected cells were collected 4 days after electroporation and total RNA was extracted using the RNeasy mini kit following manufacturer's instructions (Qiagen, Valencia, CA, USA; catalogue no. 74104). RT was carried out with 2 µL of total RNA and the Taqman RT reagents kit following manufacturer's instructions (ABI; catalogue no. N808-0234) using random hexamers as primers for RT initiation. PCR and nested PCR NS5B amplification were carried out in duplicate using the Expand High Fidelity PCR System, following manufacturer's instructions. The duplicate PCR products were pooled before being purified using the PCR purification kit (Qiagen; catalogue no. 28104), following manufacturer's instructions. Sequencing was carried out as described earlier.
Selection of mutant variants under drug pressure
For the selection of mutant variants under drug pressure using NS5B clinical isolate-derived transient replicons, 4 million cured Huh7 cells were transfected with 10 µg of in vitro-transcribed replicon RNA from clinical isolate-derived transient replicons. Reference replicons were also transfected as a control. After electroporation, cells were resuspended in 12 mL of DMEM containing 5% (v/v) fetal bovine serum and plated in 6-well plate at 600 000 cells/well. Compounds (or medium as a control) were added 24 h post-transfection at multiples of the IC50 of the respective clinical isolate populations with a final DMSO concentration of 1% (v/v) and incubated for 4 days. Sequencing of replicating replicons was performed as described earlier.
For the selection under drug pressure of the NNI-1-resistant mutant M419 in the presence of wild-type replicon Con1 (L419), a 5 µg mixture containing 10% of in vitro-transcribed M419 RNA and 90% of in vitro-transcribed L419 RNA was transfected. As a control, 5 µg of the wild-type replicon Con1 was transfected in parallel. After electroporation, cells were resuspended in 12 mL of DMEM containing 5% (v/v) fetal bovine serum and plated in 6-well plate at 600 000 cells/well. NNI-1 (or medium as a control) was added at 1×, 5× and 10×IC50 with a final DMSO concentration of 1% (v/v) and incubated for 4 days. Amplification and sequencing of replicating transient replicons were performed, as described earlier.
Replication capacity of HCV NS5B clinical isolates
The replication level of either reference strains or clinical isolate-derived transient replicons was determined as the ratio of the firefly luciferase signal at 4 days post-electroporation to the luciferase signal at 4 h post-transfection, to normalize the transfection efficiency. The replication capacity of the clinical isolate-derived replicons was expressed as their normalized replication efficiency when compared with that of the reference strain for each of the GT 1a or GT 1b, set at a value of 1.
Results
The HCV NS5B phenotypic assay mimics the patients' genetic heterogeneity and enables the determination of drug sensitivity of clinical isolates
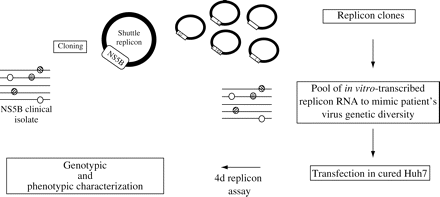
The HCV NS5B phenotypic assay enables the determination of the sensitivity of clinical isolates to polymerase inhibitors, as described in the Materials and methods section, by mimicking the intrinsic HCV genetic heterogeneity present in patients (Figure 2).
Diagram of the HCV NS5B phenotypic assay. Following PCR amplification, NS5B clinical isolates were cloned into the shuttle replicon and replicon clones were pooled to mimic the patient's virus genetic diversity. Replication capacity and drug sensitivities were evaluated after transfection of the in vitro-transcribed RNA pools into cured Huh7 cells.
The assay reproduces the genetic NS5B population present in the patient as shown by the comparison of the NS5B population sequence of the clinical isolate with the NS5B population sequence obtained after 4 days of in vitro replication (post-transfection). The NS5B region from 24 clinical isolates was sequenced either completely (amino acid residues 1–591) or partially (residues 154 and 356), and DNA sequencing data showed no major differences before and after in vitro replication, as exemplified in Figure 3. The reproducibility of the assay was evaluated by repeatedly testing the sensitivity of reference replicons from GT 1b Con1 and GT 1a H77 strains to R1479, NM107, NNI-1 and NNI-2. The mean IC50 values, standard deviations, the 95% confidence intervals and the maximum fold changes for four HCV inhibitors were obtained from between 22 and 99 independent determinations (Table 1). Most of the IC50 determinations from independent assays differed by ~2-fold with respect to the mean IC50 (with a maximum of 2.6-fold), similar to other phenotypic assays.20,21
The HCV NS5B phenotypic assay reproduces the viral genetic heterogeneity observed in patients. The sequence of the NS5B region (194–278) of the isolate population and after 4 days of in vitro replication (post-tf DNA) was compared. The box highlights the difference observed between the population sequences and post-tf DNA (X at position 254 is a mixture of amino acid residues K and R).
Reproducibility of drug sensitivity determinations
| . | . | IC50 determinations . | |||
|---|---|---|---|---|---|
| Replicon . | Statistics . | R1479 . | NM107 . | NNI-1 . | NNI-2 . |
| Con 1 | mean IC50 (μM) | 1.5 | 0.9 | 0.2 | 0.1 |
| SD | 0.6 | 0.4 | 0.1 | 0.07 | |
| na | 56 | 22 | 31 | 46 | |
| 95% CIb | 0.2 | 0.2 | 0.04 | 0.02 | |
| foldc | 2.6 | 2 | 2 | 2.3 | |
| H77 | mean IC50 (μM) | 1.7 | 0.5 | 0.5 | 0.1 |
| SD | 0.67 | 0.22 | 0.19 | 0.05 | |
| na | 99 | 41 | 50 | 55 | |
| 95% CIb | 0.2 | 0.06 | 0.06 | 0.02 | |
| foldc | 2.3 | 1.8 | 1.7 | 2.2 | |
| . | . | IC50 determinations . | |||
|---|---|---|---|---|---|
| Replicon . | Statistics . | R1479 . | NM107 . | NNI-1 . | NNI-2 . |
| Con 1 | mean IC50 (μM) | 1.5 | 0.9 | 0.2 | 0.1 |
| SD | 0.6 | 0.4 | 0.1 | 0.07 | |
| na | 56 | 22 | 31 | 46 | |
| 95% CIb | 0.2 | 0.2 | 0.04 | 0.02 | |
| foldc | 2.6 | 2 | 2 | 2.3 | |
| H77 | mean IC50 (μM) | 1.7 | 0.5 | 0.5 | 0.1 |
| SD | 0.67 | 0.22 | 0.19 | 0.05 | |
| na | 99 | 41 | 50 | 55 | |
| 95% CIb | 0.2 | 0.06 | 0.06 | 0.02 | |
| foldc | 2.3 | 1.8 | 1.7 | 2.2 | |
aRepresents the number of independent experiments.
bThe 95% confidence interval was calculated based on 2× SEM.
cMaximum fold changes relative to the mean IC50 value.
Reproducibility of drug sensitivity determinations
| . | . | IC50 determinations . | |||
|---|---|---|---|---|---|
| Replicon . | Statistics . | R1479 . | NM107 . | NNI-1 . | NNI-2 . |
| Con 1 | mean IC50 (μM) | 1.5 | 0.9 | 0.2 | 0.1 |
| SD | 0.6 | 0.4 | 0.1 | 0.07 | |
| na | 56 | 22 | 31 | 46 | |
| 95% CIb | 0.2 | 0.2 | 0.04 | 0.02 | |
| foldc | 2.6 | 2 | 2 | 2.3 | |
| H77 | mean IC50 (μM) | 1.7 | 0.5 | 0.5 | 0.1 |
| SD | 0.67 | 0.22 | 0.19 | 0.05 | |
| na | 99 | 41 | 50 | 55 | |
| 95% CIb | 0.2 | 0.06 | 0.06 | 0.02 | |
| foldc | 2.3 | 1.8 | 1.7 | 2.2 | |
| . | . | IC50 determinations . | |||
|---|---|---|---|---|---|
| Replicon . | Statistics . | R1479 . | NM107 . | NNI-1 . | NNI-2 . |
| Con 1 | mean IC50 (μM) | 1.5 | 0.9 | 0.2 | 0.1 |
| SD | 0.6 | 0.4 | 0.1 | 0.07 | |
| na | 56 | 22 | 31 | 46 | |
| 95% CIb | 0.2 | 0.2 | 0.04 | 0.02 | |
| foldc | 2.6 | 2 | 2 | 2.3 | |
| H77 | mean IC50 (μM) | 1.7 | 0.5 | 0.5 | 0.1 |
| SD | 0.67 | 0.22 | 0.19 | 0.05 | |
| na | 99 | 41 | 50 | 55 | |
| 95% CIb | 0.2 | 0.06 | 0.06 | 0.02 | |
| foldc | 2.3 | 1.8 | 1.7 | 2.2 | |
aRepresents the number of independent experiments.
bThe 95% confidence interval was calculated based on 2× SEM.
cMaximum fold changes relative to the mean IC50 value.
The variability of the phenotypic assay was also tested using NS5B clinical isolate-containing replicons generated after independent HCV RNA extraction from serum and amplification of the NS5B coding region. No significant differences in the replication capacity (data not shown) or in the levels of drug sensitivity to R1479 were observed between the two independently generated NS5B clinical isolate-containing replicons, with IC50 values within 2-fold from each other, similar to what was observed using the Con1 and H77 reference strains (Table 2).
Reproducibility of the phenotypic assay using independently generated NS5B clinical isolates containing replicons
| NS5B origina . | R1479 IC50 (μM)b . |
|---|---|
| RO-18 # 1 | 1.30 ± 0.2 (2) |
| RO-18 # 2 | 1.80 ± 0.2 (2) |
| RO-19 # 1 | 2.31 ± 0.2 (2) |
| RO-19 # 2 | 3.15 ± 0.6 (2) |
| RO-20 # 1 | 1.94 ± 0.4 (2) |
| RO-20 # 2 | 2.52 ± 0.04 (2) |
| RO-73 # 1 | 1.21 ± 0.2 (9) |
| RO-73 # 2 | 1.32 ± 0.1 (6) |
| RO-76 # 1 | 1.17 ± 0.02 (4) |
| RO-76 # 2 | 1.15 ± 0.1 (7) |
| RO-77 # 1 | 1.28 ± 0.2 (8) |
| RO-77 # 2 | 0.86 ± 0.08 (8) |
| NS5B origina . | R1479 IC50 (μM)b . |
|---|---|
| RO-18 # 1 | 1.30 ± 0.2 (2) |
| RO-18 # 2 | 1.80 ± 0.2 (2) |
| RO-19 # 1 | 2.31 ± 0.2 (2) |
| RO-19 # 2 | 3.15 ± 0.6 (2) |
| RO-20 # 1 | 1.94 ± 0.4 (2) |
| RO-20 # 2 | 2.52 ± 0.04 (2) |
| RO-73 # 1 | 1.21 ± 0.2 (9) |
| RO-73 # 2 | 1.32 ± 0.1 (6) |
| RO-76 # 1 | 1.17 ± 0.02 (4) |
| RO-76 # 2 | 1.15 ± 0.1 (7) |
| RO-77 # 1 | 1.28 ± 0.2 (8) |
| RO-77 # 2 | 0.86 ± 0.08 (8) |
a# denotes independently generated NS5B clinical isolates containing replicons.
bNumber in parentheses represents the number of independent drug-sensitive phenotypic experiments.
Reproducibility of the phenotypic assay using independently generated NS5B clinical isolates containing replicons
| NS5B origina . | R1479 IC50 (μM)b . |
|---|---|
| RO-18 # 1 | 1.30 ± 0.2 (2) |
| RO-18 # 2 | 1.80 ± 0.2 (2) |
| RO-19 # 1 | 2.31 ± 0.2 (2) |
| RO-19 # 2 | 3.15 ± 0.6 (2) |
| RO-20 # 1 | 1.94 ± 0.4 (2) |
| RO-20 # 2 | 2.52 ± 0.04 (2) |
| RO-73 # 1 | 1.21 ± 0.2 (9) |
| RO-73 # 2 | 1.32 ± 0.1 (6) |
| RO-76 # 1 | 1.17 ± 0.02 (4) |
| RO-76 # 2 | 1.15 ± 0.1 (7) |
| RO-77 # 1 | 1.28 ± 0.2 (8) |
| RO-77 # 2 | 0.86 ± 0.08 (8) |
| NS5B origina . | R1479 IC50 (μM)b . |
|---|---|
| RO-18 # 1 | 1.30 ± 0.2 (2) |
| RO-18 # 2 | 1.80 ± 0.2 (2) |
| RO-19 # 1 | 2.31 ± 0.2 (2) |
| RO-19 # 2 | 3.15 ± 0.6 (2) |
| RO-20 # 1 | 1.94 ± 0.4 (2) |
| RO-20 # 2 | 2.52 ± 0.04 (2) |
| RO-73 # 1 | 1.21 ± 0.2 (9) |
| RO-73 # 2 | 1.32 ± 0.1 (6) |
| RO-76 # 1 | 1.17 ± 0.02 (4) |
| RO-76 # 2 | 1.15 ± 0.1 (7) |
| RO-77 # 1 | 1.28 ± 0.2 (8) |
| RO-77 # 2 | 0.86 ± 0.08 (8) |
a# denotes independently generated NS5B clinical isolates containing replicons.
bNumber in parentheses represents the number of independent drug-sensitive phenotypic experiments.
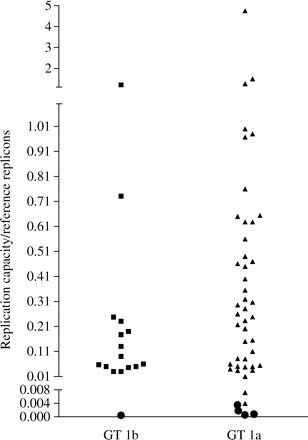
Using this phenotypic assay, a total of 63 clinical isolates from untreated patients (47 GT 1a and 16 GT 1b) were characterized. The replication capacity varied across the different clinical isolates, irrespective of the patients' viral load (data not shown), with five clinical isolates either not replicating or replicating at low levels that did not allow for an accurate IC50 determination (Figure 4). A total of 58 clinical isolates (92%) demonstrated sufficient replication levels to allow for drug sensitivity determination.
Replication capacity of NS5B clinical isolates from 63 untreated patients. Black circles indicate the clinical isolates with replication capacity that did not allow drug sensitivity determination.
Activity of HCV polymerase inhibitors against genetically diverse GT 1 clinical isolates
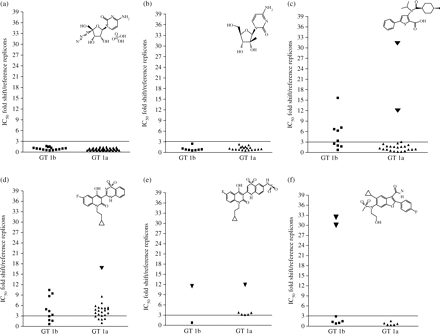
The sensitivity of GT 1 clinical isolates from untreated patients to nucleoside analogues R1479 and NM107 as well as to NNIs binding at different allosteric sites of the polymerase, thumb II (NNI-1), palm I (NNI-2 and NNI-3) and palm II (HCV-796) was determined (Figure 5). All clinical isolates were sensitive to R1479 (Figure 5a) and to NM107 (Figure 5b), within 2.5-fold of the mean IC50 (i.e. within the variability of the assay) compared with the reference strains H77 and Con1. The in vitro replication capacity of the clinical isolates did not affect the compound potency.
Sensitivity to R1479, NM107, NNI-1, NNI-2, NNI-3 and HCV-796 of untreated HCV NS5B clinical isolates indicated as IC50 fold shift, compared with reference replicons Con1 and H77. (a) A total of 58 NS5B clinical isolates (15 GT 1b and 43 GT 1a) were tested to determine their sensitivity to R1479. (b) A total of 27 NS5B clinical isolates (8 GT 1b and 19 GT 1a) were tested to determine their sensitivity to NM107. (c) Thirty-two NS5B clinical isolates (9 GT 1b and 23 GT 1a) were tested to determine their sensitivity to NNI-1 (thiophene carboxylic acid, thumb II inhibitor). Inverted black triangles indicate the GT 1a clinical isolates with M423I polymorphism (RO-67 and RO-75). (d) Thirty-three NS5B clinical isolates (10 GT 1b and 23 GT 1a) were tested to determine their sensitivity to NNI-2 (benzothiadiazine, palm I inhibitor). Inverted black triangle indicates the GT 1a clinical isolate with an H95C polymorphism (RO-58). (e) Eight NS5B clinical isolates (2 GT 1b and 6 GT 1a) were tested to determine their sensitivity to NNI-3 (thiadiazine, palm I inhibitor). Inverted black triangles indicate clinical isolates with S556G polymorphism (RO-3 and RO-38). (f) Thirteen NS5B clinical isolates (7 GT 1b and 6 GT 1a) were tested to determine their sensitivity to HCV-796 (benzofuran carboxamide, palm II inhibitor). Inverted black triangles indicate the GT 1b clinical isolates with C316N polymorphism (RO-8 and RO-13). In each graph, chemical structures of the compounds are indicated and the black line indicates the 3-fold IC50 shift compared with reference replicons.
The majority of the GT 1a isolates (21/23) were sensitive to NNI-1 (Figure 5c), whereas 2 of them (RO-67 and RO-75) demonstrated a reduced sensitivity (>12-fold compared with the reference control). Five of the nine GT 1b clinical isolates showed a decreased sensitivity to NNI-1, which ranged from 3- to 15-fold in IC50 value when compared with the reference control (Figure 5c).
In the case of palm I inhibitor NNI-2, 75% of the tested GT 1a clinical isolates (17/23) showed a decreased sensitivity to the compound that ranged from 3- to 9-fold in IC50 value versus H77 (Figure 5d), whereas one isolate had a greater reduction in sensitivity (17-fold). Six of the 10 GT 1b clinical isolates showed a decreased sensitivity to NNI-2 that ranged from 3.5- to 11-fold when compared with Con1 (Figure 5d).
Sensitivity to palm I inhibitor NNI-3 (Figure 5e) was slightly reduced (3–4-fold), compared with H77 in five of the six GT 1a clinical isolates tested and the remaining isolate showed a reduction in sensitivity of 12-fold. One GT 1b isolate was sensitive to NNI-3, whereas the other tested showed a 12-fold reduced sensitivity when compared with the reference strain Con1.
Sensitivity to HCV-796 was comparable with reference strains H77 and Con1 (Figure 5f) except in two GT 1b isolates that showed a 30-fold reduced sensitivity compared with Con1.
The presence of known drug resistance mutations or natural polymorphisms can explain reductions in drug sensitivity
To investigate the possible correlation between the observed variable sensitivity to some polymerase inhibitors and the natural genetic heterogeneity of HCV, the population sequence of the entire NS5B coding region was obtained for a total of 92 clinical isolates from treatment-naive individuals (72 GT 1a and 20 GT 1b isolates), of which 58 had been phenotypically characterized, as mentioned earlier (Figures 4 and 5). The NS5B DNA sequences were compared with their respective reference strains (H77 for GT 1a and Con1 for GT 1b isolates). No naturally occurring resistance mutations S96T, S96T/N142T or S282T, which confer resistance to nucleoside analogues R147922 and NM107,23 respectively, were observed. In contrast, although 73 of the 92 isolates (79%) showed wild-type sequence at positions related to resistance to NNIs, 19 (21%) contained natural polymorphisms at amino acid residues involved in resistance to inhibitors binding to thumb I, thumb II, palm I or palm II sites of the polymerase. Of this 21%, changes at amino acid residues that correlated with resistance to palm I-binding inhibitors were observed in 8%,7,24 2% contained mutations that conferred resistance to thumb II-binding inhibitors24 and another 2% to HCV-796.16 Amino acid substitution at residue 499 (V to A), related to resistance to thumb I inhibitors,12 was seen in 9% of the isolates and all were of GT 1b origin.
The two GT 1a clinical isolates (RO-67 and RO-75) that demonstrated reduced sensitivity to NNI-1 (Figure 5c) contained the amino acid substitution M423I in the NS5B coding region, known to confer resistance to this compound,24 either as a complete M423I substitution, which correlated with a 35-fold reduced inhibitory activity when compared with the wild-type replicon, or as a mixture of M (wild-type) and I (~30% of the mutant residue), which corresponded to a reduced inhibitory activity of 12-fold when compared with the reference control (Figure 5c and Table 3).
Amino acid substitutions that confer resistance to thumb II-, palm I- and palm II-binding inhibitors in vitro can also confer resistance in the context of clinical isolates
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Palm I inh (NNI-2) IC50 (μM) . | Palm I inh (NNI-3) IC50 (nM) . | Palm II inh (HCV-796) IC50 (nM) . | R1479 IC50 (μM) . | NM107 IC50 (μM) . |
|---|---|---|---|---|---|---|---|
| GT 1b Con1 | 0.2 ± 0.02 | 0.15 ± 0.01 | 0.9 ± 0.14 | 4.3 ± 0.7 | 1.5 ± 0.1 | 0.9 ± 0.1 | |
| RO-3 | V499A, S556G/Sb | 1.6 ± 0.22 | 0.4 ± 0.06 | 10.6 ± 2 | ND | 1.7 ± 0.2 | 0.6 ± 0.1 |
| RO-8 | C316N | 0.4 ± 0.1 | 0.5 ± 0.07 | ND | 130 ± 14 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| RO-13 | C316N | ND | 1.3 ± 0.04 | ND | 140 ± 7 | 2.3 ± 0.2 | ND |
| GT 1a H77 | 0.5 ± 0.03 | 0.1 ± 0.01 | 0.9 ± 0.09 | 5.6 ± 0.6 | 1.7 ± 0.1 | 0.5 ± 0.03 | |
| RO-58 | H95C | 0.9 ± 0.21 | 1.7 ± 0.34 | ND | ND | 0.8 ± 0.1 | ND |
| RO-38 | S556G | 0.4 ± 0.1 | 0.2 ± 0.06 | 11 ± 1.9 | ND | 1.2 ± 0.2 | ND |
| RO-67 | M423I | 17.4 ± 6.9 | 0.6 ± 0.11 | ND | ND | 0.6 ± 0.1 | ND |
| RO-75 | M423M/Ic | 6.1 ± 1.2 | 0.4 ± 0.06 | ND | ND | 1.8 ± 0.1 | ND |
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Palm I inh (NNI-2) IC50 (μM) . | Palm I inh (NNI-3) IC50 (nM) . | Palm II inh (HCV-796) IC50 (nM) . | R1479 IC50 (μM) . | NM107 IC50 (μM) . |
|---|---|---|---|---|---|---|---|
| GT 1b Con1 | 0.2 ± 0.02 | 0.15 ± 0.01 | 0.9 ± 0.14 | 4.3 ± 0.7 | 1.5 ± 0.1 | 0.9 ± 0.1 | |
| RO-3 | V499A, S556G/Sb | 1.6 ± 0.22 | 0.4 ± 0.06 | 10.6 ± 2 | ND | 1.7 ± 0.2 | 0.6 ± 0.1 |
| RO-8 | C316N | 0.4 ± 0.1 | 0.5 ± 0.07 | ND | 130 ± 14 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| RO-13 | C316N | ND | 1.3 ± 0.04 | ND | 140 ± 7 | 2.3 ± 0.2 | ND |
| GT 1a H77 | 0.5 ± 0.03 | 0.1 ± 0.01 | 0.9 ± 0.09 | 5.6 ± 0.6 | 1.7 ± 0.1 | 0.5 ± 0.03 | |
| RO-58 | H95C | 0.9 ± 0.21 | 1.7 ± 0.34 | ND | ND | 0.8 ± 0.1 | ND |
| RO-38 | S556G | 0.4 ± 0.1 | 0.2 ± 0.06 | 11 ± 1.9 | ND | 1.2 ± 0.2 | ND |
| RO-67 | M423I | 17.4 ± 6.9 | 0.6 ± 0.11 | ND | ND | 0.6 ± 0.1 | ND |
| RO-75 | M423M/Ic | 6.1 ± 1.2 | 0.4 ± 0.06 | ND | ND | 1.8 ± 0.1 | ND |
ND, not done.
aOnly amino acid substitutions related to resistance, compared with the reference strains, are indicated.
bS556G/S: G, 53%; S, 47% (determined by sequencing of the 96 replicon molecular clones present in the population).
cM423M/I: M, 70%; I, 30% (determined using the sequencing chromatograms).
Amino acid substitutions that confer resistance to thumb II-, palm I- and palm II-binding inhibitors in vitro can also confer resistance in the context of clinical isolates
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Palm I inh (NNI-2) IC50 (μM) . | Palm I inh (NNI-3) IC50 (nM) . | Palm II inh (HCV-796) IC50 (nM) . | R1479 IC50 (μM) . | NM107 IC50 (μM) . |
|---|---|---|---|---|---|---|---|
| GT 1b Con1 | 0.2 ± 0.02 | 0.15 ± 0.01 | 0.9 ± 0.14 | 4.3 ± 0.7 | 1.5 ± 0.1 | 0.9 ± 0.1 | |
| RO-3 | V499A, S556G/Sb | 1.6 ± 0.22 | 0.4 ± 0.06 | 10.6 ± 2 | ND | 1.7 ± 0.2 | 0.6 ± 0.1 |
| RO-8 | C316N | 0.4 ± 0.1 | 0.5 ± 0.07 | ND | 130 ± 14 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| RO-13 | C316N | ND | 1.3 ± 0.04 | ND | 140 ± 7 | 2.3 ± 0.2 | ND |
| GT 1a H77 | 0.5 ± 0.03 | 0.1 ± 0.01 | 0.9 ± 0.09 | 5.6 ± 0.6 | 1.7 ± 0.1 | 0.5 ± 0.03 | |
| RO-58 | H95C | 0.9 ± 0.21 | 1.7 ± 0.34 | ND | ND | 0.8 ± 0.1 | ND |
| RO-38 | S556G | 0.4 ± 0.1 | 0.2 ± 0.06 | 11 ± 1.9 | ND | 1.2 ± 0.2 | ND |
| RO-67 | M423I | 17.4 ± 6.9 | 0.6 ± 0.11 | ND | ND | 0.6 ± 0.1 | ND |
| RO-75 | M423M/Ic | 6.1 ± 1.2 | 0.4 ± 0.06 | ND | ND | 1.8 ± 0.1 | ND |
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Palm I inh (NNI-2) IC50 (μM) . | Palm I inh (NNI-3) IC50 (nM) . | Palm II inh (HCV-796) IC50 (nM) . | R1479 IC50 (μM) . | NM107 IC50 (μM) . |
|---|---|---|---|---|---|---|---|
| GT 1b Con1 | 0.2 ± 0.02 | 0.15 ± 0.01 | 0.9 ± 0.14 | 4.3 ± 0.7 | 1.5 ± 0.1 | 0.9 ± 0.1 | |
| RO-3 | V499A, S556G/Sb | 1.6 ± 0.22 | 0.4 ± 0.06 | 10.6 ± 2 | ND | 1.7 ± 0.2 | 0.6 ± 0.1 |
| RO-8 | C316N | 0.4 ± 0.1 | 0.5 ± 0.07 | ND | 130 ± 14 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| RO-13 | C316N | ND | 1.3 ± 0.04 | ND | 140 ± 7 | 2.3 ± 0.2 | ND |
| GT 1a H77 | 0.5 ± 0.03 | 0.1 ± 0.01 | 0.9 ± 0.09 | 5.6 ± 0.6 | 1.7 ± 0.1 | 0.5 ± 0.03 | |
| RO-58 | H95C | 0.9 ± 0.21 | 1.7 ± 0.34 | ND | ND | 0.8 ± 0.1 | ND |
| RO-38 | S556G | 0.4 ± 0.1 | 0.2 ± 0.06 | 11 ± 1.9 | ND | 1.2 ± 0.2 | ND |
| RO-67 | M423I | 17.4 ± 6.9 | 0.6 ± 0.11 | ND | ND | 0.6 ± 0.1 | ND |
| RO-75 | M423M/Ic | 6.1 ± 1.2 | 0.4 ± 0.06 | ND | ND | 1.8 ± 0.1 | ND |
ND, not done.
aOnly amino acid substitutions related to resistance, compared with the reference strains, are indicated.
bS556G/S: G, 53%; S, 47% (determined by sequencing of the 96 replicon molecular clones present in the population).
cM423M/I: M, 70%; I, 30% (determined using the sequencing chromatograms).
Although the five GT 1b clinical isolates that showed a 3–15-fold decreased sensitivity to NNI-1 had no known NNI-1 resistance mutations, four of them had the amino acid substitution V499A, which is in close proximity to residues in direct contact with the inhibitor and could affect its inhibitory activity.
In the case of palm I inhibitor NNI-2, the GT 1a clinical isolate (RO-58) with the lowest sensitivity to the drug (17-fold reduction; Figure 5d and Table 3) contained the amino acid substitution H95C, previously shown to confer resistance to this type of compound when the amino acid substitution is an arginine.7,24 Isolate RO-3 showed an ~3-fold decrease of sensitivity to NNI-2 despite not having any known NNI-2 resistance mutations detected by population sequencing (Table 3). The GT 1a clinical isolates with a 3–9-fold decreased sensitivity to NNI-2 had no known NNI-2 resistance mutations (Figure 5d).
Although no known NNI-2 resistance mutations have been found in the GT 1b isolates that showed a decreased sensitivity, the presence of C316N polymorphism was observed in two of them (RO-8 and RO-13; Table 3). A Con 1 replicon containing C316N showed a 3-fold reduction in sensitivity to NNI-2 (S. Rajyaguru, M. McCown and I. Najera, unpublished results), suggesting that this polymorphism (particularly in a different genetic context) could be responsible for the 3.5–8.5-fold reduced sensitivity observed in these clinical isolates.
The highest decrease in sensitivity to palm I inhibitor NNI-3 (Figure 5e and Table 3) observed in two clinical isolates (GT 1b RO-3 and GT 1a RO-38) was due to amino acid substitution at position 556 (S to G), previously shown to confer resistance to NNI-3.24 Similarly, the 30-fold reduced activity of HCV-796 observed against GT 1b clinical isolates RO-8 and RO-13 (Figure 5f and Table 3) correlated with the presence of an asparagine at amino acid residue 316.
Genetic diversity of variants within HCV quasispecies
To investigate the genetic heterogeneity of HCV quasispecies and the existence of minority mutant variants with reduced drug sensitivity, the quasispecies of 13 clinical isolates (9 GT 1a and 4 GT 1b) were analysed by DNA sequencing. Three of the selected samples contained mutations at residues 499 and 556 of the NS5B related to viral resistance to thumb I12 and palm I inhibitors, respectively,24 whereas the other 10 did not contain any resistance mutation(s) detectable by population sequencing. Amino acid alignments of the quasispecies (comprising approximately 90–100 clones) for each patient sample demonstrated different population structures in each case: within each clinical isolate, clones genetically identical to the isolate's consensus (major) sequence were detected at frequencies of 5% to 16% in 11 clinical isolates and of 2% to 4% in the other 2 isolates. The rest of the molecular replicon clones (variants) contained between 1 and 12 amino acid differences with respect to their own consensus sequence, the majority of which were unique in the population. This shows that HCV quasispecies contain a high proportion of unique variants that can allow the virus to adapt to changes in the environment.
Among the studied 1110 HCV variants, with a sequencing detection limit of minority variants present at a frequency of 1% in the quasispecies, no amino acid substitutions responsible for resistance to R1479 or NM107 were found. However, the presence of mutations that confer resistance to NNIs binding to the palm and thumb domains of the polymerase was observed within the quasispecies of untreated patients. The most prevalent mutations were those that confer resistance to palm I-binding inhibitors NNI-2 and NNI-3 observed in 11 of 13 isolates (85%) at a frequency ranging from 1% to 5% in each patient (Figure 6). Variants that contained mutations conferring resistance to the palm- and thumb-binding inhibitors co-existed in eight isolates, but were not observed together in the same molecule.
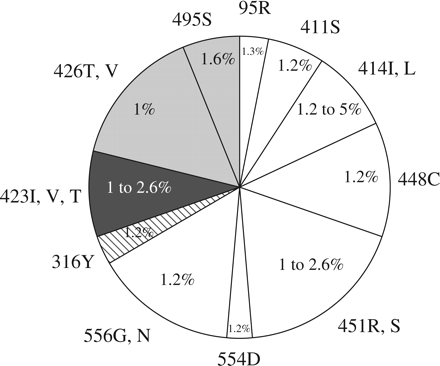
Prevalence of NNI resistance mutations among the quasispecies of untreated HCV NS5B clinical isolates. Prevalence of pre-existing NNI resistance mutations in the quasispecies of four GT 1b and nine GT 1a isolates. Amino acid substitutions found at each position are indicated. The number of molecular replicon clones in each clinical isolate containing the specified resistance mutation is indicated as a percentage. For each amino acid residue, the area of the pie chart is proportional to the number of isolates containing the specified resistance mutation in the quasispecies (resistance mutations at positions 95, 316 and 554 were seen in the quasispecies of one isolate each, 411 and 495 in two isolates each, 414 and 423 in three isolates each, 448 in four isolates, 426 and 556 in five each and 451 in six isolates). The code for pie chart represents the NNI binding sites: palm I-related amino acid residues, white; palm II-related amino acid residues, hatching; thumb I-related amino acid residues, light grey; thumb II-related amino acid residues, dark grey.
Drug sensitivity of variants within HCV quasispecies
We sought to investigate the effect that these minority variants may exert on the drug sensitivity of the clinical isolates. For this purpose, a total of 70 variants from seven clinical isolates were characterized to understand their sensitivity to inhibitors. Clones identical to their respective consensus sequence replicated at a similar or even higher level when compared with the clinical isolate population (data not shown). A qualitative correlation was also found between the number of amino acid substitutions compared with their respective consensus sequence and the variant's replication capacity: a total of 40 variants from the same seven isolates that contained three or fewer amino acid substitutions compared with their respective consensus sequence were tested and 24 were replication competent (60%). In contrast, 20 clones with four or more amino acid substitutions compared with their consensus sequence were selected from the same seven isolates and all but one were replication incompetent (data not shown).
All replicating variants were sensitive to R1479 and NM107 with a similar IC50 to their respective originating isolates (data not shown). The sensitivity to NNI-1 and NNI-2 of variants was similar to that of their respective population (data not shown), except in those cases in which known NNI resistance mutations were observed. Those variants exhibited reduced drug sensitivity when compared with their population isolates, as exemplified in Table 4. Isolate RO-65 showed only a moderate decrease in sensitivity to NNI-2. However, minor variants within its quasispecies that contained an NNI-2 resistance mutation at position 414 (methionine to isoleucine) were present at a frequency of 1.3% in the population. The variant containing M414I was isolated, tested and shown to be 15-fold less sensitive to NNI-2 when compared with the RO-65 population (Table 4). Furthermore, this variant showed similar replication capacity to the RO-65 population (data not shown).
Palm resistance mutations found at a low frequency in the quasispecies of clinical isolates confer resistance
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-2) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-3) IC50 (nM) . | Fold IC50 shift . |
|---|---|---|---|---|---|---|---|
| RO-3 population | V499A, S556G/Sb | 1.6 ± 0.22 | 1 | 0.4 ± 0.06 | 1 | 10.6 ± 2 | 1 |
| RO-3-D10 | V499A, S556G | 2.2 ± 0.02 | 1.4 | 0.4 ± 0.04 | 1 | 16.2 ± 3.1 | 1.5 |
| RO-3-D10-G556S | V499A, G556S | 2.4 ± 0.35 | 1.5 | 0.4 ± 0.03 | 1 | 0.2 ± 0.04 | 0.02 |
| RO-3-D04 | V499A, M414L | 1.4 ± 0.30 | 1.1 | >30 | >75 | ND | ND |
| RO-65 population | —c | 0.5 ± 0.12 | 1 | 0.5 ± 0.08 | 1 | ND | ND |
| RO-65-B06 | M414I | 0.3 ± 0.02 | 0.6 | 7.4 ± 0.6 | 15 | ND | ND |
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-2) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-3) IC50 (nM) . | Fold IC50 shift . |
|---|---|---|---|---|---|---|---|
| RO-3 population | V499A, S556G/Sb | 1.6 ± 0.22 | 1 | 0.4 ± 0.06 | 1 | 10.6 ± 2 | 1 |
| RO-3-D10 | V499A, S556G | 2.2 ± 0.02 | 1.4 | 0.4 ± 0.04 | 1 | 16.2 ± 3.1 | 1.5 |
| RO-3-D10-G556S | V499A, G556S | 2.4 ± 0.35 | 1.5 | 0.4 ± 0.03 | 1 | 0.2 ± 0.04 | 0.02 |
| RO-3-D04 | V499A, M414L | 1.4 ± 0.30 | 1.1 | >30 | >75 | ND | ND |
| RO-65 population | —c | 0.5 ± 0.12 | 1 | 0.5 ± 0.08 | 1 | ND | ND |
| RO-65-B06 | M414I | 0.3 ± 0.02 | 0.6 | 7.4 ± 0.6 | 15 | ND | ND |
ND, not done.
aOnly amino acid substitutions related to resistance, compared with the reference strains, are indicated.
bS556G/S: G, 53%; S, 47% and M414L: M, 95%; L, 5% (determined by sequencing of the 96 replicon molecular clones present in the population).
cRO-65 population does not contain any substitution at known resistance amino acid positions.
Palm resistance mutations found at a low frequency in the quasispecies of clinical isolates confer resistance
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-2) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-3) IC50 (nM) . | Fold IC50 shift . |
|---|---|---|---|---|---|---|---|
| RO-3 population | V499A, S556G/Sb | 1.6 ± 0.22 | 1 | 0.4 ± 0.06 | 1 | 10.6 ± 2 | 1 |
| RO-3-D10 | V499A, S556G | 2.2 ± 0.02 | 1.4 | 0.4 ± 0.04 | 1 | 16.2 ± 3.1 | 1.5 |
| RO-3-D10-G556S | V499A, G556S | 2.4 ± 0.35 | 1.5 | 0.4 ± 0.03 | 1 | 0.2 ± 0.04 | 0.02 |
| RO-3-D04 | V499A, M414L | 1.4 ± 0.30 | 1.1 | >30 | >75 | ND | ND |
| RO-65 population | —c | 0.5 ± 0.12 | 1 | 0.5 ± 0.08 | 1 | ND | ND |
| RO-65-B06 | M414I | 0.3 ± 0.02 | 0.6 | 7.4 ± 0.6 | 15 | ND | ND |
| NS5B origin . | Amino acid substitutionsa . | Thumb II inh (NNI-1) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-2) IC50 (μM) . | Fold IC50 shift . | Palm I inh (NNI-3) IC50 (nM) . | Fold IC50 shift . |
|---|---|---|---|---|---|---|---|
| RO-3 population | V499A, S556G/Sb | 1.6 ± 0.22 | 1 | 0.4 ± 0.06 | 1 | 10.6 ± 2 | 1 |
| RO-3-D10 | V499A, S556G | 2.2 ± 0.02 | 1.4 | 0.4 ± 0.04 | 1 | 16.2 ± 3.1 | 1.5 |
| RO-3-D10-G556S | V499A, G556S | 2.4 ± 0.35 | 1.5 | 0.4 ± 0.03 | 1 | 0.2 ± 0.04 | 0.02 |
| RO-3-D04 | V499A, M414L | 1.4 ± 0.30 | 1.1 | >30 | >75 | ND | ND |
| RO-65 population | —c | 0.5 ± 0.12 | 1 | 0.5 ± 0.08 | 1 | ND | ND |
| RO-65-B06 | M414I | 0.3 ± 0.02 | 0.6 | 7.4 ± 0.6 | 15 | ND | ND |
ND, not done.
aOnly amino acid substitutions related to resistance, compared with the reference strains, are indicated.
bS556G/S: G, 53%; S, 47% and M414L: M, 95%; L, 5% (determined by sequencing of the 96 replicon molecular clones present in the population).
cRO-65 population does not contain any substitution at known resistance amino acid positions.
Isolate RO-3 contained a mixture at residue 556 (~53% mutant) that correlated with a 12-fold reduced sensitivity of the viral population to NNI-3, compared with the reference strain Con1 (Table 3). To prove that substitution S556G was solely responsible for the observed reduced sensitivity to NNI-3, a single clone containing G556 (variant RO-3-D10) was selected from the isolate's population and its drug sensitivity assessed, showing reduced sensitivity to NNI-3 (Table 4). Furthermore, when the same variant was used to revert this amino acid substitution to S556, it demonstrated an 80-fold increased sensitivity to NNI-3 against the mutant variant and 50-fold against the isolate population that contained a mixture of mutant and wild-type variants (Table 4). This isolate also contained minor variant M414L (RO-3-D04) present in its quasispecies at a frequency of 5%. The sensitivity of this clinical isolate to NNI-2 was found by the phenotypic assay to be only slightly decreased compared with wild-type (3-fold). However, when a single clone RO-3-D04 (containing NNI-2 resistance mutation M414L) was selected and tested, the sensitivity to NNI-2 was shown to be decreased by >75-fold (Table 4), showing the ability of the assay to detect the effect of minor variants that may be selected upon drug pressure.
Rapid selection of minor variants upon drug pressure
Figure 6 shows that mutations that confer resistance to the palm I-binding inhibitors NNI-2 and NNI-3 were observed in 85% of the patients at a frequency ranging from 1% to 5% in each patient. Given their low frequency within the quasispecies, these mutations were only detected by extensive clonal analysis. We sought to investigate the potential effect of drug-resistant variants present at low frequency within the quasispecies upon drug selective pressure. We have shown that isolate RO-3 contained amino acid substitution S556G in ~53% of its variants. Moreover, ~5% of the variants from this isolate contained amino acid substitution M414L, which confers resistance to palm I inhibitors.24 Given the low frequency of these M414L mutant variants, this isolate's population showed only a small reduction in sensitivity to NNI-2 (∼3-fold; Table 3). Likewise, isolate RO-65 contained variants with reduced sensitivity to NNI-2 (Table 4), albeit at a low frequency in its quasispecies. To understand whether these minority variants would be selected upon drug pressure, clinical isolates RO-3 and RO-65 were transfected and incubated for 4 days in the presence of either no drug or concentrations of NNI-2 at multiples of the IC50 of the respective clinical isolates’ populations, followed by sequencing of the NS5B region (Table 4 and Figure 7). After incubation with NNI-2 at 10× IC50, variants containing 414L were selected from the RO-3 quasispecies over the consensus population (Figure 7a). The replication capacity and sensitivity to NNI-2 of one of the variants containing M414L were tested alone in the transient replicon assay (variant RO-3-D04). Interestingly, this variant showed a very poor replication capacity in the absence of drug when compared with that of the population (20-fold lower). However, in the presence of NNI-2, the replication capacity of the mutant variant increased up to an 8-fold maximum at 3 µM (~5× IC50) (data not shown), which could explain the observed rapid selection in the presence of drug observed in Figure 7(a). As shown in Table 4, this variant showed the same sensitivity to NNI-1 as the RO-3 population, but showed reduced sensitivity to NNI-2.
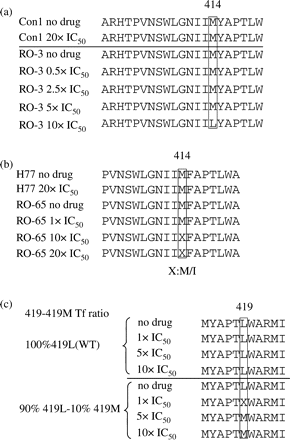
Changes in replicon population upon incubation with a thumb II-binding NNI-1 or a palm I-binding NNI-2 inhibitor. (a) RO-3 in vitro-transcribed RNA, which contained 5% of mutant M414L, was transfected and cells were incubated in the presence of concentrations of 0.5×, 2.5×, 5× and 10× IC50 of NNI-2. Cells were harvested 5 days post-transfection and total RNA was extracted. The entire NS5B region was amplified and sequenced. (b) RO-65 in vitro-transcribed RNA, which contained 1.3% of mutant M414I, was transfected and cells were incubated in the presence of concentrations of 1×, 10× and 20× IC50 of NNI-2 and processed as mentioned earlier. For both (a) and (b), only the NS5B region including residue 414 is presented here. (c) Different ratios of wild-type (419L) and mutant (419M) transient replicons were transfected and cells were incubated in the presence of 1×, 5× and 10× IC50 of NNI-1 and processed as described earlier. The NS5B region including residue 419 is presented here (X at position 419 in the 1× IC50 experiment represents a mixture M/L).
For the RO-65 isolate, a mixture of mutant 414I and wild-type was observed after incubation with 10× and 20× IC50 NNI-2 (Figure 7b). As a control, transient replicons Con1 and H77 were also incubated in the presence of NNI-1 and NNI-2 and no resistance mutation was selected in the experiment (Figure 7a and b).
To determine the frequency of a minority resistant variant in a quasispecies necessary to observe a decrease in the drug sensitivity, a wild-type GT 1b Con1 replicon (L419) and the site-directed mutant M419 were mixed at different ratios, transfected and the IC50 value for NNI-1 was determined. It should be noted that mutant L419M was used as it exhibits a similar replication capacity to the wild-type replicon. As seen in Table 5, 25% of the resistant variant needs to be present in the population to observe a reduction in sensitivity of 5-fold in a 3 day IC50 determination assay. Furthermore, sequencing of the 10% L419M/90% wild-type mixture after incubation with 1×, 5× or 10× NNI-1 IC50 shows that the variant was selected and became predominant after 4 days in the presence of 5× IC50 drug pressure (Figure 7c).
Decreased NNI-1 sensitivity correlates with the frequency of resistant mutants in replicon population
| Transfection ratio . | . | . | |
|---|---|---|---|
| L419 (wild-type) . | M419 (thumb II-binding mutant) . | IC50 (μM) . | IC50 fold shift . |
| 100% | 0% | 0.2 ± 0.01 | 1 |
| 90% | 10% | 0.4 ± 0.05 | 2 |
| 75% | 25% | 1.1 ± 0.16 | 5.5 |
| 65% | 35% | 3.9 ± 0.54 | 20 |
| 50% | 50% | 4.7 ± 0.28 | 24 |
| 0% | 100% | 7.9 ± 0.7 | 40 |
| Transfection ratio . | . | . | |
|---|---|---|---|
| L419 (wild-type) . | M419 (thumb II-binding mutant) . | IC50 (μM) . | IC50 fold shift . |
| 100% | 0% | 0.2 ± 0.01 | 1 |
| 90% | 10% | 0.4 ± 0.05 | 2 |
| 75% | 25% | 1.1 ± 0.16 | 5.5 |
| 65% | 35% | 3.9 ± 0.54 | 20 |
| 50% | 50% | 4.7 ± 0.28 | 24 |
| 0% | 100% | 7.9 ± 0.7 | 40 |
Decreased NNI-1 sensitivity correlates with the frequency of resistant mutants in replicon population
| Transfection ratio . | . | . | |
|---|---|---|---|
| L419 (wild-type) . | M419 (thumb II-binding mutant) . | IC50 (μM) . | IC50 fold shift . |
| 100% | 0% | 0.2 ± 0.01 | 1 |
| 90% | 10% | 0.4 ± 0.05 | 2 |
| 75% | 25% | 1.1 ± 0.16 | 5.5 |
| 65% | 35% | 3.9 ± 0.54 | 20 |
| 50% | 50% | 4.7 ± 0.28 | 24 |
| 0% | 100% | 7.9 ± 0.7 | 40 |
| Transfection ratio . | . | . | |
|---|---|---|---|
| L419 (wild-type) . | M419 (thumb II-binding mutant) . | IC50 (μM) . | IC50 fold shift . |
| 100% | 0% | 0.2 ± 0.01 | 1 |
| 90% | 10% | 0.4 ± 0.05 | 2 |
| 75% | 25% | 1.1 ± 0.16 | 5.5 |
| 65% | 35% | 3.9 ± 0.54 | 20 |
| 50% | 50% | 4.7 ± 0.28 | 24 |
| 0% | 100% | 7.9 ± 0.7 | 40 |
Discussion
The current standard of care provides good clinical efficacy in patients infected with HCV GTs 2 and 3, but is less efficacious in the most prevalent GT1-infected patients, thereby emphasizing the urgent need for more effective HCV-specific antiviral therapies.2,3 This has triggered considerable efforts in drug discovery with a number of HCV inhibitors currently in clinical trials, both NS3/4A protease and NS5B polymerase inhibitors. The high genetic heterogeneity of HCV, due to the error-prone nature of its RdRp, high virus production and turnover represents an opportunity for the virus to evade antiviral treatment, with every possible mutant likely present in a patient's quasispecies. The potential presence of major or minor HCV variants within a viral population carrying amino acid changes associated with reduced drug sensitivity could affect the sensitivity of untreated patients to HCV inhibitors.
In this study, we describe the development of an HCV NS5B phenotypic assay that allows for the assessment of the sensitivity of NS5B clinical isolates to polymerase inhibitors by mimicking the intrinsic HCV genetic heterogeneity present in infected patients. This approach allows for the quick determination of an isolate's drug sensitivity and, unlike other previously reported methods, avoids the need to test a large number of individual clones15 or the selection and testing of individual clones from the population25 that could potentially have significantly different genetic and phenotypic characteristics from the isolate's overall population (Table 4). As shown here, the phenotypic assay is able to detect a reduction in drug sensitivity in clinical isolates that contain natural resistance mutations at a frequency as low as 5% in the quasispecies (RO-3; see Table 3) and a significant decrease in sensitivity (12-fold IC50 shift) when mutations are present at least in 30% of the quasispecies (Table 3).
The assay described here supports the replication of >90% of genetically diverse GT 1 clinical isolates and allows for drug sensitivity determination, with a calculated variability of 2-fold (and a 2.6-fold maximum variability). Using this technology, the sensitivity of 58 clinical isolates from untreated GT 1 patients to a panel of polymerase inhibitors was determined in order to investigate the effect of HCV NS5B genetic diversity on the inhibitor's antiviral potency.
The potency of nucleoside analogues R1479 and NM107 was shown to be similar across genetically diverse clinical isolates of GT 1 origin and similar to the GT 1b Con1 and GT 1a H77 reference strains. This correlated with the lack of the in vitro-selected R1479- or NM107-resistance mutations, S96T, S96T/N142T and S282T, respectively,22,23 in the population of untreated clinical isolates. Furthermore, none of these resistance mutations was detected among the HCV quasispecies of 13 untreated patients in this study, encompassing 1110 HCV NS5B variants, within the sequencing detection limit of minority variants present at a frequency of 1% in the quasispecies. The low in vitro replication capacity of these resistant replicons of 4% for S96T, 5% for S96T-N142T22 and 20% for S282T22,23 compared with wild-type could account for the fact that they were not detected in this extensive study of the NS5B quasispecies.
In contrast, the potency of NNIs (particularly NNI-1 and NNI-2) was found to be more variable across genetically diverse clinical isolates. Indeed, using a similar type of assay, variable activity has been described for compound A-782759, chemically related to NNI-2.20 Moreover, a number of isolates that exhibited reduced drug sensitivity contained known NNI-1 or NNI-2 resistance mutations (Table 3). Several isolates were slightly less sensitive to these compounds, but did not contain any known resistance mutations (or any common mutation that could be related to this phenotype). This could be due to the intrinsic genetic diversity of HCV, by which genetically diverse clinical isolates can show variable sensitivity to inhibitors, as has been previously shown for HIV inhibitors such as enfuvirtide.26 For HCV, the existence of natural polymorphisms across different GTs has been previously shown to affect sensitivity to drugs,27,28 such as at amino acid 482 for which leucine is the wild-type consensus in GT 2a and at amino acid 423 for which isoleucine is the wild-type consensus in GT 5, suggesting that both GTs may be less sensitive to NS5B polymerase thumb II inhibitors. Likewise, other NNIs, binding to different sites of the NS5B polymerase, demonstrated a lower potency in vitro against GT 2, 3 and 4, as described previously.28 In this study, we show that the GT 1 isolates with the lowest sensitivity to NNIs contained natural amino acid polymorphisms in the NS5B coding region known to confer resistance to HCV inhibitors8,16,24 at a frequency of at least 30% in the viral population (Table 3). All but one of the clinical isolates that showed a slight decrease in sensitivity to NNI-1 and did not contain any known NNI-1 resistance mutations contained amino acid substitution V499A. Although this change has only been characterized as a thumb I resistance mutation,12 it could affect NNI-1 inhibitory activity given its proximity to the thumb II-binding site, particularly to residues Leu-497 and Arg-501, which are directly in contact with the inhibitor.24 Interestingly, GT 1a isolates showed a 3–9-fold reduced sensitivity to NNI-2. When compared with Con1, the reduced activity of compounds binding to this site of the polymerase is likely due to the effect of polymorphisms Y415F and E446Q,28 although other unknown determinants may be key for the compound binding affinity. The genetic context of each isolate can potentially affect the level of drug sensitivity: RO-3 (GT 1b) and RO-38 (GT 1a) exhibited similar levels of sensitivity to NNI-3, despite the fact that RO-38 contains only mutant G556 variants in its quasispecies, as determined by population sequencing, whereas RO-3 had a mixture of wild-type and mutant variants. Within a given genetic context (clinical isolate), however, the presence of a mixture of mutant and wild-type variants correlates with higher drug sensitivity, whereas a complete mutant population demonstrates a greater reduction in drug sensitivity, as seen in Table 4 for isolates RO-3 and RO-65 (compare sensitivity of the population versus that of the mutant or wild-type clones).
This study is the first extensive analysis of HCV NS5B quasispecies, showing that this gene exhibits a higher degree of heterogeneity than those previously reported for other regions of HCV29 or for HIV-1 protease.30 Approximately 50% of the variants were replication competent, similar to that observed for the protease gene of HIV30 and HCV.29 The replication-competent variants were found to be genetically closer to the isolate's consensus sequence, differing by three or fewer amino acid substitutions, whereas variants containing more than four amino acid substitutions were found to be replication incompetent. Given the limitation of the replicon system, we cannot exclude the possibility that incompatibility with the laboratory strain-derived replicon backbones could play a role in the replication capacity of some of these variants. Detailed molecular clonal studies showed that each quasispecies of the 13 clinical isolates had a different composition, with most of them having a major form containing those clones identical to the consensus sequence at frequencies ranging between 4% and 16%. One quasispecies (RO-30) was more heterogeneous, with only 2% of consensus clones. Variants containing one to three amino acid substitutions compared with the consensus sequence were the most abundant and, in most cases, unique. Most samples had similar viral loads (1–7 × 106 IU/mL), which did not allow for the analysis of the viral load effect on the representation of the quasispecies complexity.
The study includes a comprehensive analysis of the prevalence of mutations known to confer resistance to NS5B polymerase inhibitors among the quasispecies of untreated patients. We show here the lack of nucleoside analogue resistance mutations and the existence of NNI resistance mutations within the virus quasispecies of untreated patients. The fact that NNI-resistant variants exist in the quasispecies of HCV at a higher frequency than nucleoside analogue-resistant variants reflects the more flexible and diverse nature of the amino acid residues within the NS5B allosteric binding sites and possibly the lesser effect on replication capacity of these NNI resistance mutations. Although the presence of mutant variants at a frequency of <25% does not affect the population IC50 value significantly (Table 5) or at all (see RO-65 in Table 4), the existence of mutant variants with reduced drug sensitivity within the quasispecies of a patient will facilitate their selection upon drug selective pressure. This is shown in this study using both NS5B clinical isolates that contain a heterogeneous (wild-type and mutant) quasispecies and a site-directed mutant engineered in a Con1 transient replicon. Upon drug pressure, the quasispecies of both clinical isolates RO-3 and RO-65 change and those mutant resistant variants (present at 1.3% or 5% in the patient) become predominant in the population. Interestingly, in the case of RO-3-D04, its replication capacity increased in the presence of drug, likely facilitating its selection once under drug selective pressure. Likewise, mutant 419L when present at 10% in a viral population becomes predominant upon a 5× IC50 drug selective pressure for 4 days incubation, but not in the case of the wild-type reference replicon when under the same selection pressure (Figure 7c).
In summary, we have described an NS5B phenotypic assay that mimics the patient viral quasispecies and allows for the determination of the sensitivity to NS5B polymerase inhibitors of genetically diverse clinical isolates. The existence of NNI resistance mutations in the virus population correlated with the reduced sensitivity to NNIs observed in some clinical isolates. We also showed that resistance mutations can be present in the viral population of untreated patients at a low frequency in their quasispecies and that these variants can be readily selected upon drug pressure (Figure 7). The use of HCV phenotypic assays such as the one for the characterization of clinical isolates from clinical trials will allow the study of the levels and prevalence of resistant mutants necessary to result in clinically relevant drug resistance that may affect the efficacy of HCV inhibitors. The higher frequency of NNI resistance mutations when compared with the absence of resistance mutations to the nucleoside analogues in the HCV quasispecies of treatment-naive HCV patients suggests a potential for faster development of clinically significant resistance for non-nucleoside therapies.
Funding
This work was funded by Roche Palo Alto, LLC.
Transparency declarations
All authors are employees of Roche Palo Alto, LLC.
Acknowledgements
We thank Seth Harris for the contribution of the crystallography figure to this manuscript, Ken Brameld for NS5B modelling information, Sharon Jiang for DNA sequencing, Nixy Zutshi and her group for cell culture support and Matthew McCown for critical reading of the manuscript.