-
PDF
- Split View
-
Views
-
Cite
Cite
Rachel L Gewiss, Nathan C Law, Aileen R Helsel, Eric A Shelden, Michael D Griswold, Two distinct Sertoli cell states are regulated via germ cell crosstalk†, Biology of Reproduction, Volume 105, Issue 6, December 2021, Pages 1591–1602, https://doi.org/10.1093/biolre/ioab160
Close - Share Icon Share
Abstract
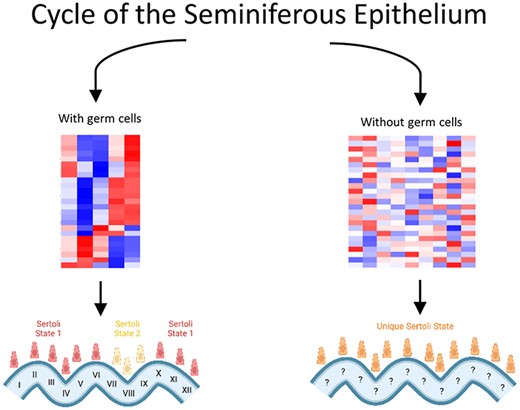
Sertoli cells are a critical component of the testis environment for their role in maintaining seminiferous tubule structure, establishing the blood-testis barrier, and nourishing maturing germ cells in a specialized niche. This study sought to uncover how Sertoli cells are regulated in the testis environment via germ cell crosstalk in the mouse. We found two major clusters of Sertoli cells as defined by their transcriptomes in Stages VII–VIII of the seminiferous epithelium and a cluster for all other stages. Additionally, we examined transcriptomes of germ cell-deficient testes and found that these existed in a state independent of either of the germ cell-sufficient clusters. Altogether, we highlight two main transcriptional states of Sertoli cells in an unperturbed testis environment, and a germ cell-deficient environment does not allow normal Sertoli cell transcriptome cycling and results in a state unique from either of those seen in Sertoli cells from a germ cell-sufficient environment.
Introduction
Complex cell-to-cell interactions within the testis have the ultimate goal of producing mature spermatozoa. While germ cells obviously play a key role in this process, somatic cells within the testis are also critical components contributing to germ cell maturation. Sertoli cells are important to provide a suitable environment for germ cells and create niches within the seminiferous epithelium that provide key nutrients and metabolites [1, 2]. Sertoli cells also offer structural support of the testis tubules so that regular testis morphology can be maintained as germ cells undergo the extreme morphological and localization changes as they mature within the tubules. Additionally, Sertoli cells secrete many proteins necessary for germ cell maturation and testis homeostasis. Metal ion binding proteins including transferrin are generated by Sertoli cells and allow iron to transit the blood-testis barrier to be accessed by germ cells [1, 3, 4]. Proteases and protease inhibitors are secreted by Sertoli cells and play a role in testis remodeling, particularly at the point of spermatid release into the tubule lumen [3, 4]. Secreted factors also include those that contribute to the tight junctions at the basement membrane and between Sertoli cells as well as hormones and growth factors necessary for spermatogenesis, including inhibin and glial cell-line-derived neurotrophic factor, as well as signals received via follicle stimulating hormone (FSH) receptors [1, 3, 4]. These factors, along with the unique shape of Sertoli cells that allows for tight-knit interactions with developing germ cells [5, 6], are critical for the normal maturation of male germ cells from spermatogonia to spermatozoa.
One major aspect of testis biology is the cycle of the seminiferous epithelium, defined by associations of germ cells that can be found within a cross-section of a tubule. There are 12 stages in the mouse cycle that have defined durations [7]. In mice, the length of one cycle in the adult is ~8.6 days [8]. The time from spermatogonial differentiation (also termed the A-to-A1 transition) to spermiation is 35 days, resulting from four full cycles. In neonatal mice undergoing the first round of spermatogonial differentiation, the cycle is somewhat shorter, such that the first cycle takes only about 6 days to complete [9, 10]. Regardless of postnatal age, retinoic acid (RA) regulates this cycle and is a trigger for spermatogonial differentiation, meiotic initiation, and spermiation [11]. RA provided by Sertoli cells is critical for spermatogonial differentiation [12, 13].
A critical question that remains is whether the presence of the cycle of the seminiferous epithelium is an intrinsic program of Sertoli cells or if Sertoli cells require crosstalk with germ cells to establish this timing, and during which developmental periods, this germ-Sertoli cell crosstalk may be necessary. Transplantation studies have shown that germ cells are able to establish niches in germ-cell ablated testes or those with transplanted Sertoli cells and show normal spermatogenesis [14–17]. However, cross-species transplantation between mice and rats have shown that the transplanted germ cells, rather than endogenous Sertoli cells, determine the timing of germ cell development and the cycle of the seminiferous epithelium. For example, rat germ cells transplanted into mouse testes adopt the timing of the rat cycle of the seminiferous epithelium, showing the importance of germ cells in regulating this timing [18]. Previous studies have found that Sertoli-germ cell crosstalk is necessary for normal function of the seminiferous epithelium [1, 19, 20–22]. We sought to determine how germ cells may be necessary for maintaining normal Sertoli cell transcriptomes throughout the cycle of the seminiferous epithelium.
To address these questions, we utilized several genomic technologies to examine Sertoli cell transcriptomes. We first used single-cell RNA-sequencing (scRNA-seq) in adult mice with testicular fractions enriched for Sertoli cells. This allowed us to see a broad picture of an unperturbed adult testis and distinguish multiple states of Sertoli cells throughout the cycle of the seminiferous epithelium. We validated these results by using WIN 18 446/RA synchronized mice and fluorescence-activated cell sorting (FACS) isolation to obtain cell populations enriched for Sertoli cells from known stages. We further assessed Sertoli cell transcriptomes in the absence of germ cells using Nanos C2HC-Type Zinc Finger 2 knock-out mice (Nanos2 KO) that present male-specific germ cell depletion and infertility [16, 22, 23]. Altogether, these data showed the cyclicity of transcript expression in Sertoli cells throughout the cycle of the seminiferous epithelium, and how these transcript patterns were altered in the absence of germ cells.
Results
Single-cell RNA-seq revealed two main Sertoli cell clusters
The most unbiased way to sample the adult Sertoli cell population while accounting for transcriptional heterogeneity is via scRNA-seq. However, previous studies have described notable limitations to this approach. Co-capture of germ and Sertoli cells is common even with FACS isolation of fluorescently labeled Sertoli cells or enrichment of germ cells [24, 25]. With this limitation in mind, we performed initial clustering of FACS isolated Sertoli cells by scRNA-seq to generate six graph-based clusters (Figure 1). Clusters were annotated for cell types using previously described germ and Sertoli cell markers [26]. Based on marker gene expression, Clusters 1 and 2 appeared highly enriched for Sertoli cells and represented ~50% of the total captured cells, a dramatic increase from the normal adult testis environment in which Sertoli cells make up ~3% of the total cell number [27] (Supplementary Figure S1).
(A) Clusters resulting from RFPxAMH-Cre single-cell sequencing of adult, unsynchronized testes. 6 major clusters were identified among the global population. (B) Heatmap representation shows the top 50 DEGs among the 6 clusters that define the population captured in library preparation. (C) Relative expression of several genes representative of both germ and somatic cells as defined by [26]. Relative abundance of germ cell genes shows that germ cells in all stages of development showed some level of co-capture with Sertoli cells.
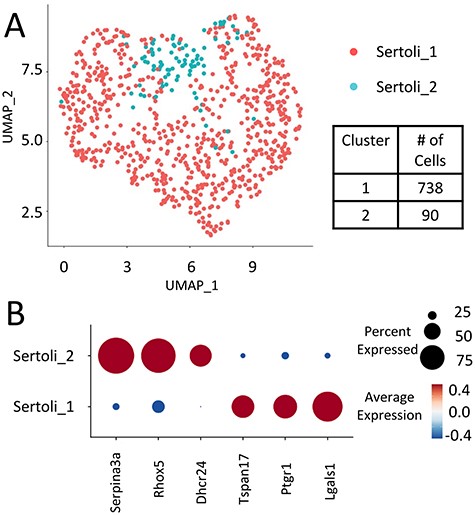
The isolated Sertoli-enriched population could be subdivided into multiple clusters. However, the differences among clusters were diminished with increasing subdivisions. For example, arbitrarily subdividing the population into eight clusters indicated that the vast majority of heterogeneity among the population was captured by just two clusters, as illustrated by a cluster dendogram (Supplementary Figure S2). Furthermore, we discovered that further subdivision of the population beyond two clusters diluted heterogeneity such that additional clusters lacked marker genes with greater than 2-fold difference compared with the remaining population. We thus used the Sertoli-enriched data divided into two clusters for further analysis (Figure 2, Supplementary File). Cluster 1 was enriched for transcripts including tetraspanin 17 (Tspan17), prostaglandin reductase 1 (Ptgr1), and galnectin 1 (Lgals1). Tspan17 transcripts have previously been shown to be highest at stages IX–XII [28], and Ptgr1 transcripts are lowest at stages VII–VIII [29]. Lgals1 is a Sertoli-enriched gene that has its highest expression values at Stages X–XII and lowest relative values at Stages VII–VIII [30]. Cluster 2 was enriched for transcripts including serpin family A member 3 (Serpina3a), reproductive homeobox 5 (Rhox5), and 24-dehydrocholesterol reductase (Dhcr24). Rhox5 has previously been shown to have its highest transcript levels in stages VII–VIII [31]. Thus, it appeared that the two clusters of Sertoli cells were divided into one group that was enriched in cells from stages VII–VIII (Cluster 2) and another group that was enriched for cells outside of stages VII–VIII (Cluster 1).
(A) Sertoli clusters (Clusters 1 and 2 from full dataset) re-divided into two main Sertoli clusters based on differential transcriptomes from the Sertoli-enriched clusters. (B) Select stage-specific genes’ representation between Sertoli clusters 1 and 2. Serpina3a, Rhox5, and Dhcr24 were highly enriched in Cluster 2 cells, while Tspan17, Ptgr1, and Lgals1 were highly enriched in Cluster 1 cells.
Adult Sertoli cell cycling in synchronized testes
Previous work has given an overview of stage-specific testis transcripts in the mouse using a RiboTag model followed by microarray [32]. We expanded this information by performing RNA-sequencing on mice with synchronized spermatogenesis. Mice were synchronized according to the previously described WIN 18 446/RA protocol [33]. To accomplish synchronization, RA synthesis is blocked such that a pool of undifferentiated spermatogonia builds up within the seminiferous epithelium before a bolus of RA is injected to stimulate the vast majority of germ cells to synchronously undergo spermatogonial differentiation. This procedure has been shown to keep germ cells developing synchronously and remain within ~3 adjacent stages of the seminiferous epithelium for several weeks following RA injection [33]. We used mice with a tdTomato reporter (denoted as RFP) crossed with AMH-Cre mice and collected several animals each day from 42 to 50 days postinjection (dpi) to represent a full cycle of the seminiferous epithelium following the first wave of spermatogenesis. One testis from each animal was used to stage the cycle of the seminiferous epithelium, and these were subsequently clustered into five groups centering around Stages I–III (Group A), IV–VI (Group B), VII–VIII (Group C), IX–X (Group D), and XI–XII (Group E).
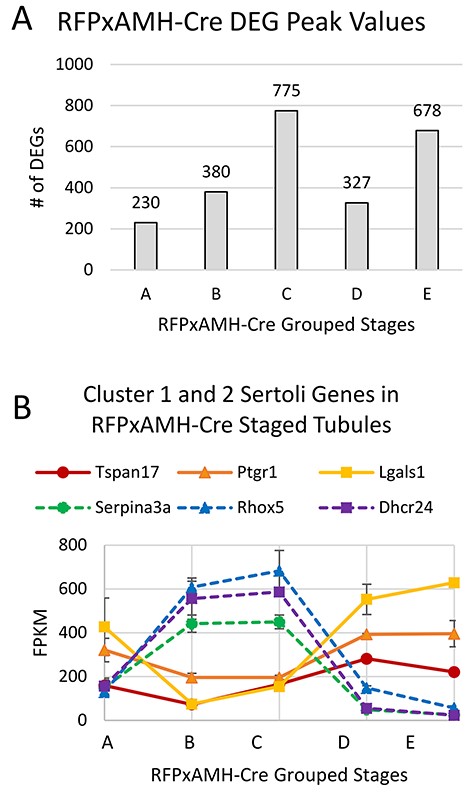
RNA-sequencing revealed 2390 genes that were differentially expressed in at least one group of stages (Supplementary File). Over 32% of these differentially expressed genes (DEGs) had peak values in Group C cells (Figure 3A). We examined some of the highly expressed and highly differential genes between the two single-cell clusters (Figure 3B). Tspan17, Ptgr1, and Lgals1, which were all enriched in Cluster 1 of our scRNA-seq data, were all lowest in Sertoli cells from Groups B and C and peaked in cells from Group D and E. Conversely, Serpina3a, Rhox5, and Dhcr24 all showed peak transcript levels in Groups B and C, and lowest transcript levels were observed in Groups D and E. This corroborated what was inferred from the scRNA-seq data and supported that Cluster 1 cells are primarily those outside of stages VII–VIII, while Cluster 2 cells comprise those that came from stage VII–VIII tubules.
(A) RFPxAMH-Cre isolated Sertoli cells had 2390 total DEGs across the cycle of the seminiferous epithelium. Graph shows the number of grouped stages of the seminiferous epithelium where DEGs displayed maximal transcript numbers. (B) Expression of key Cluster 1 or 2 genes identified from scRNA-seq in stage-synchronized RFPxAMH-Cre testes. Consistent with the single-cell data, Tspan17, Ptgr1, and Lgals1 all showed their lowest transcript levels in Group B and C tubules (stages IV–VI and VII–VIII; solid lines). Serpina3a, Rhox5, and Dhcr24 all had peak transcript levels in Group C tubules (stages VII–VIII; dashed lines).
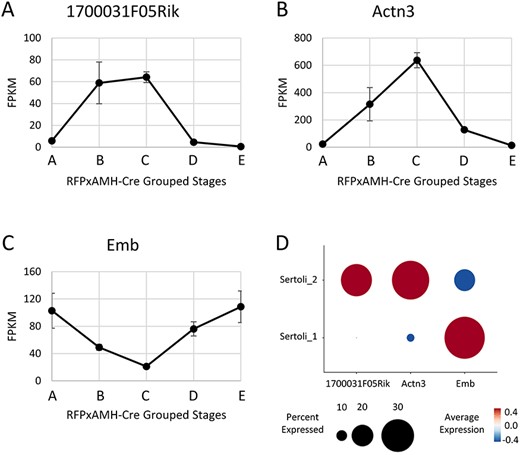
Additionally, we examined some of the transcripts with the greatest changes across the cycle of the seminiferous epithelium without defined functions in the testis (Figure 4). One such transcript was 1700031F05Rik, a doublesex and mab-3 related transcription factor (Dmrt)-like gene located on the X chromosome. While Dmrt genes on autosomes have been well-characterized with functions in male germ cell fate and maintenance, those on the X chromosome have been less well-studied [34, 35]. We found peak levels of this transcript in cells from Stage VII–VIII tubules in our bulk RNA-seq, as well as a significantly greater percentage of Cluster 2 cells containing this transcript compared with Cluster 1 cells in our scRNA-seq data. While no known function for this transcript has been identified, its higher levels in Stages VII–VIII suggest that it may play a role in the RA response occurring at these stages, although there is currently no evidence of direct involvement. Several other genes involved in cytoskeletal dynamics and cell adhesion including actinin alpha 3 (Actn3) and embigin (Emb) have previously been implicated in Sertoli cell function [36]. We found peak levels of Actn3 and lowest levels of Emb in Stage VII–VIII tubules, also reflected in our scRNA-seq. These data suggest the regulation of these genes to be important for Sertoli-germ interactions, particularly when spermiation occurs at Stage VIII.
Expression of transcripts potentially involved with regulation in Stages VII–VIII (1700031F05Rik (A) and Actn3 (B)) or outside of Stages VII–VIII (Emb (C)). Data represents the average of two replicates, error bars show ±SEM. (D) Single-cell sequencing expression of 170003F05Rik, Actn3, and Emb. Data used was from the two isolated Sertoli cell clusters.
RA pathway transcripts in isolated Sertoli cells
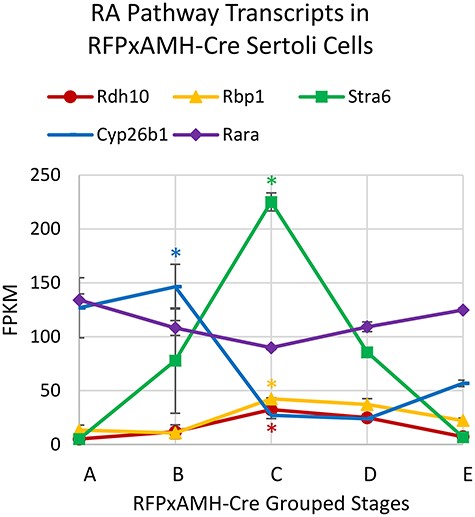
We further validated our results by examining expression differences in Sertoli cell genes known to show changes in their steady-state mRNA levels over the cycle of the seminiferous epithelium. RA is the main trigger for the cycle of the seminiferous epithelium [37], so we examined some transcripts involved in the RA pathway in the testis that are expressed in a stage-dependent manner (Figure 5). Consistent with previous data, peak levels of retinol dehydrogenase 10 (Rdh10) were detected in Group C tubules (stages VII–VIII) [37]. This corresponds with the high levels of RA during those stages and reflects the capacity of Sertoli cells with this enzyme to respond to the RA pulse [37, 38]. Another component of the RA pathway, retinol binding protein 1 (Rbp1), has been detected in Stage X–XI Sertoli cells and are abundant in Leydig cells [39]. In our data, we found this transcript to have its highest levels in cells from Groups C and D. Stimulated by RA 6 (Stra6) is important for the uptake of retinol into target cells [40]. As expected, we observed peak levels of Stra6 transcripts in Group C tubules. Cytochrome p450 family 26 subfamily B member 1 (CYP26B1) is important for clearing RA from the testis following the RA pulse, and previous reports have shown that ablation of this enzyme from the seminiferous epithelium results in abnormal testis morphology and subfertility [41]. We found high levels of Cyp26b1 in Group B cells just preceding the time of the RA pulse. This result is likely due to the need to degrade RA outside of Stages VII–VIII to prevent premature spermatogenic events, as spermatogonia in stages II–VI are susceptible to differentiation when an RA pulse is administered [37]. Furthermore, RA receptor alpha (RARα), the main RA receptor within Sertoli cells, has been previously shown to not cycle during the cycle of the seminiferous epithelium [39]. Consistent with this, we did not find Rara transcripts to vary significantly in any group of stages across the cycle of the seminiferous epithelium. Overall, we found that our isolated Sertoli cells reflected previous observations for RA pathway transcript abundance during and outside of the RA pulse.
RA pathway gene mRNA levels across the length of the cycle of the seminiferous epithelium for RFPxAMH-Cre grouped stages. Data represents the average of two replicates, error bars show ±SEM. Peak transcript levels shown by asterisks when log2 fold-change >1, P < 0.05.
Adult Sertoli cells do not cycle in the absence of germ cells
Previous evidence has shown that both Sertoli and germ cells show cyclic gene expression changes as they progress through the cycle of the seminiferous epithelium [32, 42]. To explore the role of germ cell-Sertoli cell interactions in cyclic gene expression, we utilized a Nanos2 KO mouse line in which adult males lack a germline [16, 23]. We treated these mice with WIN 18 446/RA and aged them to 42–50 dpi to mimic the timing used for the RFPxAMH-Cre bulk RNA-seq experiments. These mice could not be staged according to the cycle of the seminiferous epithelium since those distinctions are based upon germ cell morphology. Instead, we collected testes from several mice at each day postinjection within the given time frame and compared each timepoint to the others to determine transcriptome changes that were occurring over the normal length of the cycle of the seminiferous epithelium. Notably, germ cell markers including stimulated by RA gene 8 (Stra8), protamines 1 and 2 (Prm1/2), and transition proteins 1 and 2 (Tnp1/2) had expression values <1 FPKM, indicating a robust elimination of maturing germ cells.
Over the 9-day period, there were only 306 genes that were differentially expressed in at least one of the days compared with all others (Supplementary File). This was in striking contrast to the 2390 DEGs seen in the RFPxAMH-Cre dataset, suggesting that cycling of transcripts in Sertoli cells is very muted or absent in a germ cell-deficient environment. The changes in the Nanos2 KO animals were generally of lower magnitude, as 449 RFPxAMH-Cre transcripts had peak values over twice the average expression, compared with only 26 transcripts with twice the average expression in Nanos2 KO cells. Furthermore, pathway analysis of the 26 Nanos2 KO transcripts did not reveal these transcripts to be enriched for any testis or fertility terms. To better understand how DEGs in a germ cell-sufficient testis were affected by the absence of germ cells, we compared how the top cycling DEGs of RFPxAMH-Cre animals were expressed in the Nanos2 KO (Figure 6). While it was not possible to ascribe stages to Nanos2 KO testes, it was clear that the majority of cycling genes in the RFPxAMH-Cre animals did not show similar expression patterns across the time period covering one cycle of the seminiferous epithelium. Genes which were shown to be indicators of Cluster 1 and Cluster 2 cells in the scRNA-seq data were not expressed in the same cyclical manner for the stages in which they showed highest/lowest expression in a germ cell-sufficient testis (Figure 7A). We also examined some RA pathway genes that showed differential expression in the RFPxAMH-Cre cells (Figure 7B). Rdh10, Stra6, and Rara did not show differential levels in any of the days postinjection in the Nanos2 KO mice. Rbp1 had significantly higher transcript levels at 50 dpi comparatively to other days in the Nanos2 KO mice, but the trend in expression was not cyclical. Similarly, Cyp26b1 had significantly higher transcript levels at 42 dpi but then was relatively constant for the remainder of the cycle. Taken together, the greatly reduced number of DEGs in Nanos2 KO animals relative to RFPxAMH-Cre animals and the abnormal expression of top DEGs seen in a germ cell-containing testis indicate that the lack of germ cells greatly affects the gene expression profiles of Sertoli cells in a germ cell-depleted testis. Identifying which stage-specific transcripts do not show differential levels in the Nanos2 KO mice can help to indicate which gene functions are necessary for proper cycling properties of Sertoli cells in addition to those already described.
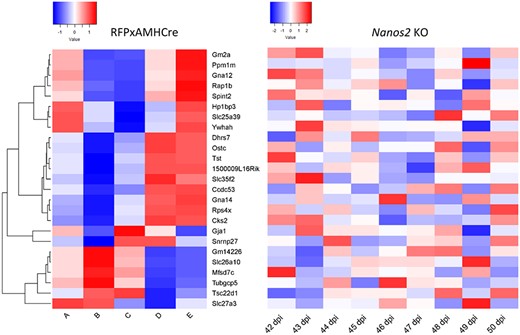
Top 25 RFPxAMH-Cre DEGs expression in RFPxAMH-Cre animals (left) and Nanos2 KO animals (right). Nanos2 KO animals lack normal cycling of transcripts seen in wild-type Sertoli cells. Color represents Z-score deviation across all samples within a condition (RFPxAMH-Cre or Nanos2 KO), with red showing greater than average representation and blue showing lower than average representation across the population.
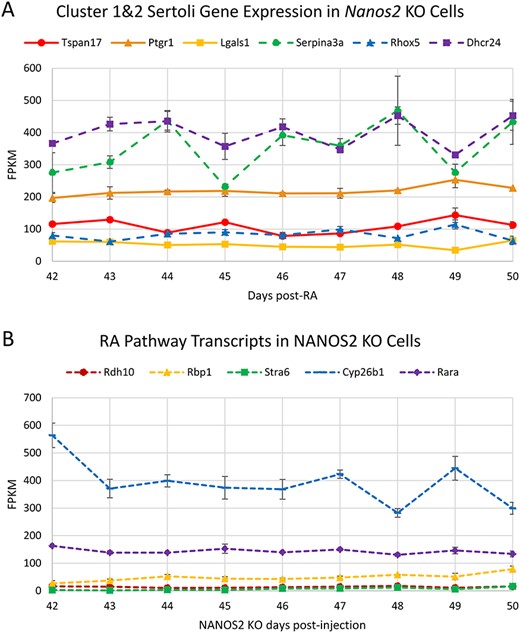
(A) Transcripts defining Sertoli cell Clusters 1 and 2 in the scRNA-seq clusters RNA-seq values in Nanos2 KO testes, and (B) RA-responsive gene mRNA levels across the length of the cycle of the seminiferous epithelium in NANOS2 KO mice 42–50 days post-RA injection. Data represents the average of two replicates, error bars show ±SEM. Peak transcript levels shown by asterisks when log2 fold-change >1, P < 0.05.
Discussion
We have previously shown how spermatogonial transcriptomes change during development from undifferentiated through B spermatogonia [43]. Here, we sought to understand how Sertoli cell transcriptomes change during the length of one cycle of the seminiferous epithelium to gain a better understanding of the somatic components of testis transcript regulation. Our data described here utilized sensitive scRNA-seq and a comprehensive deep sequencing approach to highlight the somatic contributions to the cycling of the seminiferous epithelium. We were able to assess what occurs in a synchronized system by grouping stages of tubules and using cell sorting to isolate Sertoli cells taken from an environment containing developing germ cells. This allowed us to assign stages to cell clusters that were generated through scRNA-seq. Additionally, we were able to address the question of whether normal cycling of the seminiferous epithelium occurs in the absence of germ cells by using a Nanos2 KO mouse line.
Sertoli cells show two major transcriptome states
Previous data have shown that RA causes significant changes in both testis architecture and cellular transcriptomes [11, 37]. Particularly in the germ cell-sufficient environment that we examined with RFPxAMH-Cre mice, we found that there were many transcriptome changes that occurred in tubules centered around stages VII–VIII (Group C). While Group C tubules contained the greatest number of transcripts induced to peak levels, there were also a significant number of transcripts that had greatly reduced levels in Group C tubules. Particularly in the adult mouse, Sertoli cells and developing germ cells are tightly associated via desmosome-like junctions and apical ectoplasmic specializations [44, 45] and are difficult to parse apart using traditional methods to generate single-cell suspensions. Indeed, our experiments and previous scRNA-seq studies have shown Sertoli and germ cell transcripts intermixed when supposedly only one of these cell types was being isolated [24, 25].
As an additional rigorous step, we also used an in silico negative selection approach to remove cells with notable expression of germ, Leydig, and peritubular myoid cell markers from the two main clusters of enriched Sertoli cells as previously described [26]. These remaining cells had a high purity of single Sertoli cells but also clustered similarly into two main groups with similar expression of the marker transcripts for Sertoli cells in Stages VII–VIII and all other stages (Supplementary Figure S3). Differential transcripts for these two clusters are listed in Supplementary File. Due to the greatly reduced number of cells, these data were not used for further analysis but complemented the results obtained from the larger batch of Sertoli cells.
The idea for two main Sertoli cell clusters, one for those in Stages VII–VIII where RA levels are high, and the other for cells in all other stages of the seminiferous epithelium, has been previously proposed [45]. Study of mRNA levels in the rat for transferrin, transferrin receptor, Fsh, Fshr, androgen receptor, androgen binding protein, and the Sertoli-specific isoform of Rara helped to define this hypothesis as either peak or lowest levels of these transcripts were found to occur close to Stage VIII. While levels of these transcripts have either been refined or shown to be different in the mouse, the idea stands that many transcripts that show cyclic expression in Sertoli cells have either minimal or maximal levels around Stage VIII.
Worth noting is our clustering practice for Groups A–E of RFPxAMH-Cre isolated Sertoli cells. While the majority of tubules within a cross-section displayed the stages for the group to which they were assigned, there were some tubules within the testes that were outside of those groupings. For example, Group C tubules were centered around Stages VII–VIII, although these sections sometimes contained tubules that were in surrounding stages (i.e., VI or XI). Testes from 7 to 11 mice were pooled together for each replicate for each group of stages; thus, there is some inherent heterogeneity in the groups. Additionally, the group of cells ascribed to Stages VII–VIII (Group C or single-cell cluster 2) highlights the changes that occur due to the RA pulse. The RA response is not notable in Stage VII tubules, as STRA8 is not present in germ cells of that stage [37]. However, the levels of RA in the testis remain high in Stage IX [37]; thus, the RA response begins in Group C Sertoli cells but continues to progress in cells assigned to Group D. This is shown in some of the highlighted transcripts involved in the RA pathway that have peak levels in Group C cells but remain relatively elevated in Group D before a sharp decline in Group E.
Sertoli cells require interactions with germ cells for normal cycling
Our comparison of the RFPxAMH-Cre Sertoli cells and the Nanos2 KO cells suggests that Sertoli cells are unable to properly cycle in the absence of germ cells. We discovered a greatly reduced number of DEGs in the Nanos2 KO adults and a lesser degree of transcriptome changes over the 9-day collection period. Additionally, some of the key RA pathway genes that presumably help regulate transcriptome cycling were not expressed with the same patterns, particularly Rdh10 and Stra6. The comparison of the RFPxAMH-Cre and Nanos2 KO cells suggests that Sertoli cell cycling is not maintained in the absence of germ cells.
While Sertoli function appears to be dysregulated in the absence of germ cells, previous evidence suggests that this is not a permanent state. Indeed, one of the advantages of the Nanos2 KO animals is that they can receive spermatogonial stem cell transplants and achieve normal spermatogenesis derived from the donor cells [16, 23]. Because of this ability to restart spermatogenesis, the loss of germ cells via a Nanos2 KO does not render these animals permanently sterile if donor germ cells are introduced. This is promising for use in livestock with desirable traits to be easily spread through a population as well as for men who are infertile but still have sufficient testicular architecture [46, 47]. Our data suggest Sertoli cells in a germ cell-deficient environment rest at a unique ground state that does not correspond to specific stages of the cycle of the seminiferous epithelium. This is in accordance with previous data suggesting that Sertoli cells do not rest in a particular stage of the seminiferous epithelium when vitamin A is depleted, although readministration of vitamin A leads to Sertoli cell transcriptomes similar to those normally seen in Stages VII–VIII [48]. Further study is necessary to determine how the RA pulse is regenerated when germ cells are introduced into a germ cell-deficient environment to resume normal spermatogenesis and return to the cycling of two major Sertoli cell states.
Materials and methods
Animals
All animal experiments were approved by the Washington State University Animal Care and use Committees and were conducted in accordance with the principles for the care and use of research animals of the National Institutes of Health. Nanos2 KO animals were produced on a CD1 background and were a gift from Dr Jon Oatley [23]. Female Nanos2 KO animals were bred with male Nanos2 heterozygotes to generate Nanos2 KO male progeny used for experimentation. RFP (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; Jackson Laboratory, Bar Harbor, ME, USA) homozygous animals were bred with AMH-Cre homozygous animals (C57BL/6J) [49] to produce RFPxAMH-Cre heterozygous progeny that were used for experimentation. Animals were housed in a humidity- and temperature-controlled environment with food and water provided ad libitum. At the time of tissue collection, mice were euthanized via carbon dioxide asphyxiation followed by either decapitation (less than 21 dpp) or cervical dislocation (21 dpp and older).
Mouse genotyping
Tail clips were taken from neonatal mice to identify genotypes prior to euthanasia. These tail clips were lysed in 75 μl of 25 mM NaOH, 200 μM EDTA (pH 12) in a 95°C heat block for 1 h, with manual dissociation at 30 min; 75-μl neutralization buffer (40 mM Tris–HCl, pH 5) was then added before DNA was used for genotyping reactions. Nanos2 mice were genotyped for presence or absence of the Nanos2 gene using primers specific to Nanos2 (forward 5′-TAT AAG GGG GTG TGC CTC CTC-3′, reverse 5′-TGG GTT GTG GGT GAC AGG TA-3′). Genotyping reaction and thermocycler conditions were followed according to [23]. The wild-type band is present at 559 bp, and the mutant band is present at 191 bp. RFPxAMH-Cre mice were genotyped for the presence of RFP (WT forward 5′-AAG GAG CTG CAG TGG ACT A-3′, WT reverse 5′-CCG AAA ATC TGT GGG AAG T-3′, RFP forward 5′-CTG TTC CTG TAC GGC ATG G-3′, RFP reverse 5′-GGC ATT AAA GCA GCG TAT CC-3′) and AMH-Cre (WT forward 5′-CTA TCG TGG ATC AGC GAC CTT C-3′, WT reverse 5′-CAC GGG AAC GAA ATG AAC ACC-3′, AMH-Cre forward 5′-GCG GTC TGG CAG TAA AAA CTA TC-3′, AMH-Cre reverse 5′-GTGAAA CAG CAT TGC TGT CAC TT-3′). The RFP WT band is present at 397 bp, RFP-positive band is at 196 bp, AMH-Cre WT band is at 180 bp, and the AMH-Cre-positive band is at 100 bp. RFP mice were purchased from the Jackson Laboratory, Bar Harbor, ME, USA (stock no. 007914) and AMH-Cre mice were a gift from Dr Marie-Claude Hofman.
WIN 18 446/RA treatments and testis collection
Spermatogenesis synchronization was performed as previously described [50]. Briefly, mice were treated with 100-mg/kg body weight WIN 18 446 suspended in 1% gum tragacanth every 24 h for 7 days starting at 2 dpp. At 9 dpp, mice were injected intraperitoneally with 200-μg RA diluted in 10-μl dimethyl sulfoxide (DMSO). Nanos2 KO mice were aged out 42–50 days postinjection (dpi) following WIN 18 446/RA treatment. Both testes were snap-frozen on dry ice to be used for RNA isolation. RFPxAMH-Cre adults were also aged out 42–50 days postinjection. One testis was collected for cell sorting to be used for RNA isolation, while the other testis was put in Bouin’s fixative [71% saturated picric acid (21 g/L), 24% formaldehyde, 5% glacial acetic acid] for 4 h at room temperature.
Cell sorting
Cell sorting was performed on RFPxAMH-Cre animals to collect RFP-positive Sertoli cells for sequencing. To create a single cell suspension, testes were detunicated and added to 10 mL of collagenase (1 mg/mL, X7H9763A, Worthington Biochemical, Lakewood, NJ, USA) and 1-mL DNase (7 mg/mL, 9003-98-9, Sigma, St. Louis, MO, USA) diluted in 1× HBSS (14 175 145, ThermoFischer, Waltham, MA, USA) in a 50-mL tube. Tubules were dissociated by pipetting up and down once with a 10-mL serological pipet then swirling the tube several times before incubating in a 37°C water bath for 3 min. After incubation, testes were swirled again vigorously and incubated for 3 more 3-min periods. Tubes were then placed on ice for 1 min to collect tubules to the bottom and solution was removed. Tubules were then rinsed with 10-mL 1× HBSS, swirled, set on ice for 1 min, and supernatant removed for a total of three rinses; 9-mL 0.25% trypsin and 1-mL DNase were added and pipetted up and down 10 times with a p1000 pipette to dissociate the tubules and were then put in a 37°C water bath for 5 min. Another 1 mL of DNase was added, pipetted up and down, and incubated for an additional five times; 10% of the total volume of FBS was then added and cells were put through at 40-μM cell strainer that was prerinsed with 1-mL HBSS. Cells were spun down at 600 × g for 7 min at 4°C. Cell counts were estimated using a hemocytometer and resuspended at 4 million cells/mL in 10% DNase in 1× DPBS (14 190 235, ThermoFisher, Waltham, MA, USA). FACS was performed with an SH800 machine (Sony Biotechnology, San Jose, CA, USA) to obtain RFP-positive Sertoli cells; 78% RFP-positive cells were obtained for scRNA-seq, and ~95% RFP-positive cells were collected for bulk RNA-seq experiments (representative purity graph shown in Supplementary Figure S4). Collected cells were spun down post-sorting at 600 × g for 7 min at 4°C. Supernatant was removed and cells were resuspended in 100-μL Trizol (Invitrogen, Waltham, MA, USA) and stored at −20°C until RNA extraction.
Staging RFPxAMH-Cre testes
One testis per animal was fixed in Bouin’s then taken through a graded ethanol series to dehydrate the tissue prior to embedding in paraffin wax. Tissues were cut with a microtome into 4-μM sections and placed on Superfrost Plus slides (ThermoFisher, Waltham, MA, USA). To clarify stages of the seminiferous epithelium, immunohistochemistry was performed as previously described [51] with anti-STRA8 produced in-house [37] at a 1:500 dilution and horse anti-rabbit secondary antibody at 1:2000 dilution. Stages were then determined via characterization of the mouse seminiferous epithelium [52–54]. Synchrony was tight with ~3–4 stages present per animal. Due to the need for larger mRNA quantities, testes from multiple mice were combined into grouped stages: Stages I–III (Group A), IV–VI (Group B), VII–VIII (Group C), IX–X (Group D), and XI–XII (Group E). Two replicates of each group were used for RNA-seq.
RNA sample preparation and sequencing
To isolate total RNA, the TRIzol RNA isolation procedure was followed according to the manufacturer’s instructions. Resulting RNA pellets were resuspended in 20-μL warm nuclease-free water and stored at −80°C until further use. mRNA was enriched via an NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490S, New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions; 10 ng of mRNA was used to make barcoded libraries using the Ion Total RNA-seq Kit V2 (Life Technologies, Carlsbad, CA, USA). Resulting libraries were quantified and pooled to 75 pM before loading onto Ion P1 semiconductor chips using an Ion Chef and sequenced on an Ion Proton sequencer. Reads were mapped to the Mus musculus genome v10 and expression values were calculated using a Bowtie/TopHat/Cufflinks pipeline incorporated into the Ion Torrent Suite v.5.0.5 [55, 56].
Bulk RNA-Seq data analysis
Differential gene expression was determined using the R DESeq2 package [57]. DEGs were defined as those that had a log2 fold-change ≥1 and FDR ≤ 0.05. Heat maps were generated through R version 4.0.2 gplots package [58]. Transcripts used for heat map construction were vetted to only include those also present in the scRNA-seq dataset enriched for two Sertoli cell clusters.
Single-cell RNA sequencing and analysis
Fluorescently labeled cells were isolated from adult Amh-CreTg;RFP+/− testes via cell sorting. Cells from triplicate males were pooled at equal proportions and loaded into a Chromium Controller (10× Genomics, Inc., Pleasanton, CA, USA). Single-cell cDNA libraries were generated using v2 chemistry according to the manufacturers protocol (10× Genomics, Inc.) and sequenced via Illumina HiSeq 4000 (Genomics and Cell Characterization Core Facility, University of Oregon). Raw data were processed, demultiplexed, and aligned to the 10-mm mouse transcriptome using the 10× Genomics Cell Ranger pipeline (v2.1.0) with “force-cells” set to 2500 for accurate identification of cells based on anticipated capture.
After raw data processing, the resulting gene-cell matrix was analyzed further in R software using the Seurat package (v3.2.2). Transcriptomes were excluded if the percent of reads mapping to the mitochondrial genome was greater than 25%. Gene expression data were then normalized, scaled (“vars.to.regress = nCount_RNA”), and variable genes identified using the default parameters in Seurat. Dimensional reduction was then performed via principal component (PC) analysis and the number of PCs (40 PCs) was selected based on statistical significance (P > 0.05) using the “JackStraw” procedure. Individual cells were projected onto UMAP two-dimensional representations using the selected PCs. Graph-based clustering via Louvain algorithm with the “FindClusters” function (resolution = 0.35) generated six clusters. DEGs between the six clusters were calculated using “FindAllMarkers” under default parameters.
Based on enrichment of Sertoli cell-specific genes and reduced germ cell-specific genes, clusters 1 and 2 were isolated from the remaining population for further analysis. The subsetted gene expression matrix was reassembled in Seurat repeating the procedures above, including normalization, scaling, variable gene identification, dimensional reduction (6 PCs), and clustering (resolution = 0.12). Reclustering generated largely overlapping clusters compared with the original clusters used for subsetting. Genes differentially expressed between the two resulting clusters was calculated with “FindAllMarkers” (min.pct = 0.1). For the negative selection of Sertoli cells by removing other testicular cell types, cells were removed if they had expression >0.1 of the normalized read counts for the following genes (obtained from [26]): Ddx4, Dazl, Id4, Stra8, Kit, Meiob, Meioc, Sycp3, Dmc1, Ccnb1, Adad2, Cyp2a12, Klk1b3, Cby3, Ssxb2, Speer4e, Acot10, Adam29, Pecam1, Vwf, Myh11, Cxc3r1, and Adgre1. This reduced the cell count of 1188 cells of the two isolated Sertoli cell clusters down to 492 cells (41.4%).
Data availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [59] and are accessible through GEO Series accession number GSE166563. Additionally, interactive visualization of scRNA-seq data is available at simplesinglecell.org/griswold/.
Author contributions
Project conceptualization, MG, RG, and AH; data acquisition, RG and AH; manuscript writing and editing, RG, NL, ES, and MG; analysis, RG, NL, ES, and MG; figures, RG, ES, and NL; funding acquisition, MG.
Acknowledgments
The authors thank Dr. John Amory for supplying WIN 18 446, the Washington State University Laboratory for Biotechnology and Bioanalysis for sample sequencing and initial data processing, the Center for Reproductive Biology FACS Core for assistance with cell sorting, and Traci Topping for technical assistance.
Conflicts of interest
The authors declare no conflicts of interest.
Grant Support: This work was supported by NIH grant R01 HD10808 (to MDG).



![(A) Clusters resulting from RFPxAMH-Cre single-cell sequencing of adult, unsynchronized testes. 6 major clusters were identified among the global population. (B) Heatmap representation shows the top 50 DEGs among the 6 clusters that define the population captured in library preparation. (C) Relative expression of several genes representative of both germ and somatic cells as defined by [26]. Relative abundance of germ cell genes shows that germ cells in all stages of development showed some level of co-capture with Sertoli cells.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/biolreprod/105/6/10.1093_biolre_ioab160/1/m_ioab160f1.jpeg?Expires=1716668601&Signature=n4U8PaisPpR~qc6GJ-JUJ6hMaaIx9n1XcULyOQ8iEcnnxHKTokumTXAfDiEz812TH4RusDnWWyQOsgzQDv2G-YSrtj-JdtarAv4MTLG9LZX3o7cJi7Uu6jVtnonxKvWIWBfYz2ZakDP05oQ9wb28WEclCF3AFwAtrb7Ei2rcOR4XRVYpYPcAbJGSxOtZw18e6UOmJFJqyZGqfoj2N7T1IyP5R0-JqVDH5Fnvd3KQi-jmpYrpvHiGaVnqf2sPPs~OEZC64hxnTGRIo0l3X-GxIs-CpUOTn1qqms9nvVK~TXRCvwBRAHK~vOZeccdDu-Ub2dZOtavrsQB1rcRlNW-Z1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)