-
PDF
- Split View
-
Views
-
Cite
Cite
Atsushi Hakura, Hiroyasu Shimada, Madoka Nakajima, Hajime Sui, Sachiko Kitamoto, Satoshi Suzuki, Tetsuo Satoh, Salmonella/human S9 mutagenicity test: a collaborative study with 58 compounds, Mutagenesis, Volume 20, Issue 3, May 2005, Pages 217–228, https://doi.org/10.1093/mutage/gei029
Close - Share Icon Share
Abstract
A large and extensive body of data on the use of human liver S9 fractions in the Salmonella mutagenicity test (Ames test) is presented; the data were obtained from a collaborative study by JEMS/BMS (Bacterial Mutagenicity Test Study Group) members and the Human and Animal Bridging Research Organization (HAB). In this study, the mutagenicity of 58 chemicals, many of which were judged to be human carcinogens by the IARC, was determined by the Ames test (the pre-incubation method at 37°C for 20 min) in the presence of a selected human liver S9 fraction with a high drug-metabolic activity or a pooled human liver S9 fraction with a moderate drug-metabolic activity. For reference, mutagenicity was also examined in the presence of a phenobarbital/5,6-benzoflavone-pretreated rat liver S9 fraction, which is normally used in mutagenicity testing systems. The bacterial test strains consisted of Salmonella typhimurium TA100, TA98 or YG7108. The data indicated that the mutagenicity of chemicals in the rat and human liver S9 fractions varied considerably, depending on the chemicals in question. In addition, a large inter-individual diversity in the mutagenic response to mutagens, depending on the chemical structures of the mutagens, was also demonstrated using two selected human S9 fractions. Most of the mutagens tested in this study (75%; 36 out of 48 compounds that were judged to be mutagenic in at least one S9 fraction) were less mutagenic in the presence of the two human S9 fractions than in the presence of the rat S9 fraction. On the other hand, the other compounds (25%), including some aromatic amines and nitrosamines, showed a more potent mutagenicity in the presence of either one of the two human S9 fractions than in the presence of the rat S9 fraction. These data strongly suggest that the use of human liver S9 fraction in mutagenicity testing systems may be useful for a better understanding of the mutagenic effects of chemicals on humans.
Introduction
The Salmonella mutagenicity test (Ames test) is used worldwide as an initial screening to determine the mutagenic potential of new chemicals for hazard identification and in documents for the registration or acceptance of new chemicals by regulatory agencies (1,2). Many carcinogens remain inactive until they are enzymatically transformed to an electrophilic species that is capable of covalently binding to DNA leading to mutation; thus, metabolic activation is considered to be a critical step in mutation (3–6). S9 fractions prepared from the livers of rats pretreated with phenobarbital (PB)/5,6-benzoflavone (BF) or Aroclor 1254 to induce drug metabolizing enzyme activity are very useful for mutagenicity screening systems, since they effectively bioactivate promutagens to mutagens and are also convenient and cost-effective (1,2,7,8). In addition, consistent responses to test substances may be important for regulatory decision-making.
The use of human S9 fractions in the Ames test is starting to attract attention because of recent advances in the acquisition of human materials for research in addition to the value of these human S9 fractions in evaluating mutagenicity in humans. However, available data on the mutagenicity of chemicals in the presence of human S9 fractions, particularly pooled S9 fractions prepared from multiple donor tissues, is limited (3,9–29). One of the reasons for this limited amount of data was probably the difficulty in acquiring human materials, which were not commercially available. Another reason may be that much attention has been paid to the sensitive detection of the mutagenicity (hazard identification) of chemicals using rodent S9 fractions with a high drug-metabolic activity, such as those prepared from rats treated with PB/BF or Aroclor 1254 or from hamsters (7,8,30,31). The purpose of this collaborative study was to accumulate a large and extensive body of data on the use of human S9 fractions in the Ames test to examine the differences in the mutagenicity of chemicals in the presence of rat and human S9 fractions.
Our previous study (28) suggested that a selected human liver S9 fraction with a high drug-metabolic activity, such as the Human S9 (HLS-014) fraction (to be abbreviated as HLS-014), as well as a pooled human liver S9 fraction (to be abbreviated as pooled S9) with a moderate drug-metabolic activity may be useful in mutagenicity testing systems as a measure of evaluating inter-individual variability in mutagenic response to chemicals among humans. In a previous study, HLS-014 showed the most potent mutagenic activity to three known procarcinogens among human liver S9 fractions prepared from 18 donated liver tissues stocked in the Biomedical Research Institute. Accordingly, the same human S9 fractions (HLS-014 and pooled S9) were used in this collaborative study. A PB/BF-induced rat liver S9 fraction (to be abbreviated as rat S9) was also used as a reference. In the present study, we conducted the Ames test in the presence of each S9 fraction for 58 chemicals, many of which have been identified as human carcinogens by the IARC (IARC Monographs home page, available at http://monographs.iarc.fr/monoeval/grlist.html). Hence, the diversity in the mutagenic activities of the test compounds in the presence of human and rat S9 fractions is discussed with special attention to the chemical structures of the test compounds. The advantages and disadvantages of using human S9 fractions in mutagenicity testing systems are also discussed.
Materials and methods
Participating laboratories
The laboratories participating in this collaborative study are listed in Table I.
Participants in the collaborative study
| Lab No. . | Laboratory . | Investigators . |
|---|---|---|
| 1 | Biosafety Research Center, Foods, Drugs and Pesticides (An-Pyo Center) | Madoka Nakajima, Jin Tanakaa, Yumie Haruta |
| 2 | Canon Inc. | Fukue Andoha, Takako Kobayashi, Hiroko Hayashi |
| 3 | Daiichi Pharmaceutical Co., Ltd | Miki Miuraa, Miyuki Igarashi, Satoru Itoh, Shiho Nakayama, Yayoi Hayasaki |
| 4 | Eisai Co., Ltd | Atsushi Hakuraa, Shigeki Sawada, Tadakazu Sugihara, Yuji Hori, Kanako Uchida |
| 5 | Environmental Biological Life Science Research Center, Inc. (BILIS) | Noriko Hashimotoa, Tagahiko Morinaga |
| 6 | Food and Drug Safety Center | Hajime Suia, Hiroko Komeya, Takumi Hara |
| 7 | Fuji Biomedix Co., Ltd | Takeshi Sakataa, Masaomi Indo |
| 8 | Fuji Photo Film Co., Ltd | Masaharu Fujitaa, Masako Yanagawa, Fujiko Soga |
| 9 | General Testing Research Institute of Japan Oilstuff Inspectors' Corporation | Masato Nakamuraa |
| 10 | Hoyu Co., Ltd | Toshitaka Osakia |
| 11 | Japan Food Research Laboratories | Yukiko Kaneuji, Mitsuko Takahashi, Kayo Matsushita, Hidetaka Satoa |
| 12 | Kissei Pharmaceutical Co., Ltd | Kazuo Kobayashia, Kayoko Takashima, Yukari Okino, Shigenari Ozawa |
| 13 | Lion Co., Ltd | Mitsuru Haresakua, Yasuhito Yamamoto |
| 14 | Nihon Bayer Agrochem K. K. | Kiyoto Satakea |
| 15 | Nikken Chemical Co., Ltd | Yuko Saitoa |
| 16 | Nippon Experimental Medical Research Institute Co., Ltd | Haruki Inouea, Golam Sarwar, Matsumi Takano |
| 17 | Nippon Menard Cosmetic Co., Ltd | Hajime Kojimaa |
| 18 | Nippon Shinyaku Co., Ltd | Yasuhiro Yamashitaa, Hideyuki Tamura |
| 19 | Nissan Chemical Industries, Ltd | Idumi Ogawa, Rie Kubotaa |
| 20 | Shin Nippon Biomedical Laboratories, Ltd | Kazuhiko Saigo, Naohiko Torigoea |
| 21 | Sumitomo Chemical Co., Ltd | Sachiko Kitamotoa, Mika Ota |
| 22 | Taisho Pharmaceutical Co., Ltd | Kiyohiro Hashimotoa, Koh-ichi Ohsawa |
| 23 | Toyama Chemical Co., Ltd | Chikako Yoshidaa, Harumi Tonoyama, Harumi Araki |
| 24 | Toyo Ink MFG. Co., Ltd | Ikuko Tsuchikoa |
| 25 | Tsumura & Co. | Makoto Katamia, Toshihiro Kammoto |
| Lab No. . | Laboratory . | Investigators . |
|---|---|---|
| 1 | Biosafety Research Center, Foods, Drugs and Pesticides (An-Pyo Center) | Madoka Nakajima, Jin Tanakaa, Yumie Haruta |
| 2 | Canon Inc. | Fukue Andoha, Takako Kobayashi, Hiroko Hayashi |
| 3 | Daiichi Pharmaceutical Co., Ltd | Miki Miuraa, Miyuki Igarashi, Satoru Itoh, Shiho Nakayama, Yayoi Hayasaki |
| 4 | Eisai Co., Ltd | Atsushi Hakuraa, Shigeki Sawada, Tadakazu Sugihara, Yuji Hori, Kanako Uchida |
| 5 | Environmental Biological Life Science Research Center, Inc. (BILIS) | Noriko Hashimotoa, Tagahiko Morinaga |
| 6 | Food and Drug Safety Center | Hajime Suia, Hiroko Komeya, Takumi Hara |
| 7 | Fuji Biomedix Co., Ltd | Takeshi Sakataa, Masaomi Indo |
| 8 | Fuji Photo Film Co., Ltd | Masaharu Fujitaa, Masako Yanagawa, Fujiko Soga |
| 9 | General Testing Research Institute of Japan Oilstuff Inspectors' Corporation | Masato Nakamuraa |
| 10 | Hoyu Co., Ltd | Toshitaka Osakia |
| 11 | Japan Food Research Laboratories | Yukiko Kaneuji, Mitsuko Takahashi, Kayo Matsushita, Hidetaka Satoa |
| 12 | Kissei Pharmaceutical Co., Ltd | Kazuo Kobayashia, Kayoko Takashima, Yukari Okino, Shigenari Ozawa |
| 13 | Lion Co., Ltd | Mitsuru Haresakua, Yasuhito Yamamoto |
| 14 | Nihon Bayer Agrochem K. K. | Kiyoto Satakea |
| 15 | Nikken Chemical Co., Ltd | Yuko Saitoa |
| 16 | Nippon Experimental Medical Research Institute Co., Ltd | Haruki Inouea, Golam Sarwar, Matsumi Takano |
| 17 | Nippon Menard Cosmetic Co., Ltd | Hajime Kojimaa |
| 18 | Nippon Shinyaku Co., Ltd | Yasuhiro Yamashitaa, Hideyuki Tamura |
| 19 | Nissan Chemical Industries, Ltd | Idumi Ogawa, Rie Kubotaa |
| 20 | Shin Nippon Biomedical Laboratories, Ltd | Kazuhiko Saigo, Naohiko Torigoea |
| 21 | Sumitomo Chemical Co., Ltd | Sachiko Kitamotoa, Mika Ota |
| 22 | Taisho Pharmaceutical Co., Ltd | Kiyohiro Hashimotoa, Koh-ichi Ohsawa |
| 23 | Toyama Chemical Co., Ltd | Chikako Yoshidaa, Harumi Tonoyama, Harumi Araki |
| 24 | Toyo Ink MFG. Co., Ltd | Ikuko Tsuchikoa |
| 25 | Tsumura & Co. | Makoto Katamia, Toshihiro Kammoto |
Corresponding investigator of each laboratory.
Participants in the collaborative study
| Lab No. . | Laboratory . | Investigators . |
|---|---|---|
| 1 | Biosafety Research Center, Foods, Drugs and Pesticides (An-Pyo Center) | Madoka Nakajima, Jin Tanakaa, Yumie Haruta |
| 2 | Canon Inc. | Fukue Andoha, Takako Kobayashi, Hiroko Hayashi |
| 3 | Daiichi Pharmaceutical Co., Ltd | Miki Miuraa, Miyuki Igarashi, Satoru Itoh, Shiho Nakayama, Yayoi Hayasaki |
| 4 | Eisai Co., Ltd | Atsushi Hakuraa, Shigeki Sawada, Tadakazu Sugihara, Yuji Hori, Kanako Uchida |
| 5 | Environmental Biological Life Science Research Center, Inc. (BILIS) | Noriko Hashimotoa, Tagahiko Morinaga |
| 6 | Food and Drug Safety Center | Hajime Suia, Hiroko Komeya, Takumi Hara |
| 7 | Fuji Biomedix Co., Ltd | Takeshi Sakataa, Masaomi Indo |
| 8 | Fuji Photo Film Co., Ltd | Masaharu Fujitaa, Masako Yanagawa, Fujiko Soga |
| 9 | General Testing Research Institute of Japan Oilstuff Inspectors' Corporation | Masato Nakamuraa |
| 10 | Hoyu Co., Ltd | Toshitaka Osakia |
| 11 | Japan Food Research Laboratories | Yukiko Kaneuji, Mitsuko Takahashi, Kayo Matsushita, Hidetaka Satoa |
| 12 | Kissei Pharmaceutical Co., Ltd | Kazuo Kobayashia, Kayoko Takashima, Yukari Okino, Shigenari Ozawa |
| 13 | Lion Co., Ltd | Mitsuru Haresakua, Yasuhito Yamamoto |
| 14 | Nihon Bayer Agrochem K. K. | Kiyoto Satakea |
| 15 | Nikken Chemical Co., Ltd | Yuko Saitoa |
| 16 | Nippon Experimental Medical Research Institute Co., Ltd | Haruki Inouea, Golam Sarwar, Matsumi Takano |
| 17 | Nippon Menard Cosmetic Co., Ltd | Hajime Kojimaa |
| 18 | Nippon Shinyaku Co., Ltd | Yasuhiro Yamashitaa, Hideyuki Tamura |
| 19 | Nissan Chemical Industries, Ltd | Idumi Ogawa, Rie Kubotaa |
| 20 | Shin Nippon Biomedical Laboratories, Ltd | Kazuhiko Saigo, Naohiko Torigoea |
| 21 | Sumitomo Chemical Co., Ltd | Sachiko Kitamotoa, Mika Ota |
| 22 | Taisho Pharmaceutical Co., Ltd | Kiyohiro Hashimotoa, Koh-ichi Ohsawa |
| 23 | Toyama Chemical Co., Ltd | Chikako Yoshidaa, Harumi Tonoyama, Harumi Araki |
| 24 | Toyo Ink MFG. Co., Ltd | Ikuko Tsuchikoa |
| 25 | Tsumura & Co. | Makoto Katamia, Toshihiro Kammoto |
| Lab No. . | Laboratory . | Investigators . |
|---|---|---|
| 1 | Biosafety Research Center, Foods, Drugs and Pesticides (An-Pyo Center) | Madoka Nakajima, Jin Tanakaa, Yumie Haruta |
| 2 | Canon Inc. | Fukue Andoha, Takako Kobayashi, Hiroko Hayashi |
| 3 | Daiichi Pharmaceutical Co., Ltd | Miki Miuraa, Miyuki Igarashi, Satoru Itoh, Shiho Nakayama, Yayoi Hayasaki |
| 4 | Eisai Co., Ltd | Atsushi Hakuraa, Shigeki Sawada, Tadakazu Sugihara, Yuji Hori, Kanako Uchida |
| 5 | Environmental Biological Life Science Research Center, Inc. (BILIS) | Noriko Hashimotoa, Tagahiko Morinaga |
| 6 | Food and Drug Safety Center | Hajime Suia, Hiroko Komeya, Takumi Hara |
| 7 | Fuji Biomedix Co., Ltd | Takeshi Sakataa, Masaomi Indo |
| 8 | Fuji Photo Film Co., Ltd | Masaharu Fujitaa, Masako Yanagawa, Fujiko Soga |
| 9 | General Testing Research Institute of Japan Oilstuff Inspectors' Corporation | Masato Nakamuraa |
| 10 | Hoyu Co., Ltd | Toshitaka Osakia |
| 11 | Japan Food Research Laboratories | Yukiko Kaneuji, Mitsuko Takahashi, Kayo Matsushita, Hidetaka Satoa |
| 12 | Kissei Pharmaceutical Co., Ltd | Kazuo Kobayashia, Kayoko Takashima, Yukari Okino, Shigenari Ozawa |
| 13 | Lion Co., Ltd | Mitsuru Haresakua, Yasuhito Yamamoto |
| 14 | Nihon Bayer Agrochem K. K. | Kiyoto Satakea |
| 15 | Nikken Chemical Co., Ltd | Yuko Saitoa |
| 16 | Nippon Experimental Medical Research Institute Co., Ltd | Haruki Inouea, Golam Sarwar, Matsumi Takano |
| 17 | Nippon Menard Cosmetic Co., Ltd | Hajime Kojimaa |
| 18 | Nippon Shinyaku Co., Ltd | Yasuhiro Yamashitaa, Hideyuki Tamura |
| 19 | Nissan Chemical Industries, Ltd | Idumi Ogawa, Rie Kubotaa |
| 20 | Shin Nippon Biomedical Laboratories, Ltd | Kazuhiko Saigo, Naohiko Torigoea |
| 21 | Sumitomo Chemical Co., Ltd | Sachiko Kitamotoa, Mika Ota |
| 22 | Taisho Pharmaceutical Co., Ltd | Kiyohiro Hashimotoa, Koh-ichi Ohsawa |
| 23 | Toyama Chemical Co., Ltd | Chikako Yoshidaa, Harumi Tonoyama, Harumi Araki |
| 24 | Toyo Ink MFG. Co., Ltd | Ikuko Tsuchikoa |
| 25 | Tsumura & Co. | Makoto Katamia, Toshihiro Kammoto |
Corresponding investigator of each laboratory.
Chemicals
Table II lists the name, CAS no., source and purity of the test chemicals that were used as well as the laboratory (see corresponding number in Table I) where the assay was performed.
Chemical mutagens assayed in this study
| Chemical no. . | Chemical name . | CAS no. . | Source . | Purity (%) . | Assay lab no. . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polycyclic hydrocarbons | ||||||||||
| [1] | Phenanthrene | 85-01-8 | Tokyo Kasei | >98 | 7 | |||||
| [2] | Pyrene | 129-00-0 | Tokyo Kasei | >97 | 17 | |||||
| [3] | Chrysene | 218-01-9 | Tokyo Kasei | 99 | 10 | |||||
| [4] | Benz[a]anthracene | 56-55-3 | Tokyo Kasei | >99 | 25 | |||||
| [5] | Dibenz[a,c]anthracene | 215-58-7 | Tokyo Kasei | >99 | 15 | |||||
| [6] | Dibenz[a,h]anthracene | 53-70-3 | Tokyo Kasei | >95 | 17 | |||||
| [7] | Benzo[a]pyrene | 50-32-8 | Tokyo Kasei | >95 | 4 | |||||
| [8] | 7,12-Dimethylbenz[a]anthracene | 57-97-6 | Nacalai | 98 | 20 | |||||
| [9] | 3-Methylcholanthrene | 56-49-5 | Aldrich | 98 | 8 | |||||
| Heterocyclic hydrocarbons | ||||||||||
| [10] | Quinoline | 91-22-5 | Sigma | 96 | 24 | |||||
| [11] | 1,7-Phenanthroline | 230-46-6 | Aldrich | 100 | 14 | |||||
| [12] | 4-Methylquinoline (Lepidine) | 491-35-0 | Sigma | 99 | 24 | |||||
| [13] | 7,9-Dimethylbenz[c]acridine | 963-89-3 | Aldrich | 92 | 10 | |||||
| [14] | Aflatoxin B1 | 1162-65-8 | Wako | >97 | 4 | |||||
| [15] | Quercetin | 117-39-5 | Tokyo Kasei | 100 | 4 | |||||
| Aromatic amines | ||||||||||
| [16] | 2,4-Diaminotoluene | 95-80-7 | Tokyo Kasei | 98 | 5 | |||||
| [17] | 3,3′-Dimethylbenzidine (o-Tolidine) | 119-93-7 | Wako | >97 | 21 | |||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 91-94-1 | Sigma | >99 | 8 | |||||
| [19] | 4,4′-Methylenedianiline | 101-77-9 | Tokyo Kasei | 100 | 12 | |||||
| [20] | Auramine (Basic Yellow 2) | 2465-27-2 | Tokyo Kasei | 81 | 22 | |||||
| [21] | 1-Naphthylamine HCl | 552-46-5 | Tokyo Kasei | >97 | 22 | |||||
| [22] | 2-Aminofluorene | 153-78-6 | Wako | >97 | 13 | |||||
| [23] | 2-Acetylaminofluorene | 53-96-3 | Tokyo Kasei | >98 | 4 | |||||
| [24] | 2-Aminoanthracene | 613-13-8 | Wako | 95 | 4 | |||||
| [25] | 6-Aminochrysene | 2642-98-0 | Aldrich | 97 | 4 | |||||
| [26] | 1-Aminopyrene | 1606-67-3 | Tokyo Kasei | >95 | 19 | |||||
| Heterocyclic amines | ||||||||||
| [27] | PhIP HCl | 105650-23-5 | Wako | 99 | 1 | |||||
| [28] | Trp-P-2 acetate | 72254-58-1 | Wako | 97 | 25 | |||||
| [29] | MeAαC acetate | 68006-83-7a | Wako | 99 | 14 | |||||
| [30] | IQ | 76180-96-6 | Wako | >99 | 7 | |||||
| [31] | MeIQx | 77500-04-0 | Wako | 99 | 1 | |||||
| Aromatic nitro compounds | ||||||||||
| [32] | Chloramphenicol | 56-75-7 | Sigma | 98 | 18 | |||||
| [33] | Metronidazole | 443-48-1 | Tokyo Kasei | 100 | 18 | |||||
| [34] | 2,4-Dinitrotoluene | 121-14-2 | Wako | 99 | 23 | |||||
| [35] | 2-Nitrofluorene | 607-57-8 | Tokyo Kasei | 99 | 11 | |||||
| [36] | 1-Nitronaphthalene | 86-57-7 | Kanto Chemical | 98 | 11 | |||||
| [37] | Azathioprine | 446-86-6 | Sigma | 98 | 3 | |||||
| [38] | AF-2 | 3688-53-7 | Wako | 99 | 23 | |||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | 56-57-5 | Tokyo Kasei | 98 | 5 | |||||
| [40] | 1-Nitropyrene | 5522-43-0 | Aldrich | 99 | 3 | |||||
| Azo dyes | ||||||||||
| [41] | Azobenzene | 103-33-3 | Tokyo Kasei | 100 | 2 | |||||
| [42] | 4-Aminoazobenzene | 60-09-3 | Tokyo Kasei | 100 | 2 | |||||
| [43] | o-Aminoazotoluene | 97-56-3 | Tokyo Kasei | >95 | 13 | |||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 60-11-7 | Tokyo Kasei | GRb | 2 | |||||
| [45] | Congo red (Direct Red 28) | 573-58-0 | Wako | GR | 9 | |||||
| [46] | Trypan blue | 72-57-1 | Aldrich | 96 | 21 | |||||
| [46′] | Trypan blue (crude) | 72-57-1 | Nacalai | GR | 21 | |||||
| Nitrosamines | ||||||||||
| [47] | N-Nitrosodimethylamine | 62-75-9 | Tokyo Kasei | 100 | 4 | |||||
| [48] | N-Nitrosodiethylamine | 55-18-5 | Tokyo Kasei | >99 | 4 | |||||
| [49] | N-Nitroso-di-n-propylamine | 621-64-7 | Tokyo Kasei | 100 | 4 | |||||
| [50] | N-Nitroso-di-n-butylamine | 924-16-3 | Tokyo Kasei | 100 | 4 | |||||
| [51] | N-Nitrosomorpholine | 59-89-2 | Tokyo Kasei | 100 | 4 | |||||
| Vinyl compounds | ||||||||||
| [52] | Acrylonitrile | 107-13-1 | Tokyo Kasei | >99 | 15 | |||||
| [53] | Acrylamide | 79-06-1 | Nacalai | 100 | 20 | |||||
| [54] | Styrene | 100-42-5 | Tokyo Kasei | >99 | 9 | |||||
| [55] | Safrole | 94-59-7 | Tokyo Kasei | >96 | 19 | |||||
| Miscellaneous nitrogen compounds | ||||||||||
| [56] | Hydrazine 2HCl | 5341-61-7 | Tokyo Kasei | >98 | 6 | |||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 306-37-6 | Tokyo Kasei | >98 | 6 | |||||
| [58] | Dacarbazine | 4342-03-4 | Sigma | 98 | 12 | |||||
| [59] | Sodium nitrite | 7632-00-0 | Wako | 99 | 16 | |||||
| [60] | Sodium nitrate | 7631-99-4 | Wako | 99 | 16 | |||||
| Chemical no. . | Chemical name . | CAS no. . | Source . | Purity (%) . | Assay lab no. . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polycyclic hydrocarbons | ||||||||||
| [1] | Phenanthrene | 85-01-8 | Tokyo Kasei | >98 | 7 | |||||
| [2] | Pyrene | 129-00-0 | Tokyo Kasei | >97 | 17 | |||||
| [3] | Chrysene | 218-01-9 | Tokyo Kasei | 99 | 10 | |||||
| [4] | Benz[a]anthracene | 56-55-3 | Tokyo Kasei | >99 | 25 | |||||
| [5] | Dibenz[a,c]anthracene | 215-58-7 | Tokyo Kasei | >99 | 15 | |||||
| [6] | Dibenz[a,h]anthracene | 53-70-3 | Tokyo Kasei | >95 | 17 | |||||
| [7] | Benzo[a]pyrene | 50-32-8 | Tokyo Kasei | >95 | 4 | |||||
| [8] | 7,12-Dimethylbenz[a]anthracene | 57-97-6 | Nacalai | 98 | 20 | |||||
| [9] | 3-Methylcholanthrene | 56-49-5 | Aldrich | 98 | 8 | |||||
| Heterocyclic hydrocarbons | ||||||||||
| [10] | Quinoline | 91-22-5 | Sigma | 96 | 24 | |||||
| [11] | 1,7-Phenanthroline | 230-46-6 | Aldrich | 100 | 14 | |||||
| [12] | 4-Methylquinoline (Lepidine) | 491-35-0 | Sigma | 99 | 24 | |||||
| [13] | 7,9-Dimethylbenz[c]acridine | 963-89-3 | Aldrich | 92 | 10 | |||||
| [14] | Aflatoxin B1 | 1162-65-8 | Wako | >97 | 4 | |||||
| [15] | Quercetin | 117-39-5 | Tokyo Kasei | 100 | 4 | |||||
| Aromatic amines | ||||||||||
| [16] | 2,4-Diaminotoluene | 95-80-7 | Tokyo Kasei | 98 | 5 | |||||
| [17] | 3,3′-Dimethylbenzidine (o-Tolidine) | 119-93-7 | Wako | >97 | 21 | |||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 91-94-1 | Sigma | >99 | 8 | |||||
| [19] | 4,4′-Methylenedianiline | 101-77-9 | Tokyo Kasei | 100 | 12 | |||||
| [20] | Auramine (Basic Yellow 2) | 2465-27-2 | Tokyo Kasei | 81 | 22 | |||||
| [21] | 1-Naphthylamine HCl | 552-46-5 | Tokyo Kasei | >97 | 22 | |||||
| [22] | 2-Aminofluorene | 153-78-6 | Wako | >97 | 13 | |||||
| [23] | 2-Acetylaminofluorene | 53-96-3 | Tokyo Kasei | >98 | 4 | |||||
| [24] | 2-Aminoanthracene | 613-13-8 | Wako | 95 | 4 | |||||
| [25] | 6-Aminochrysene | 2642-98-0 | Aldrich | 97 | 4 | |||||
| [26] | 1-Aminopyrene | 1606-67-3 | Tokyo Kasei | >95 | 19 | |||||
| Heterocyclic amines | ||||||||||
| [27] | PhIP HCl | 105650-23-5 | Wako | 99 | 1 | |||||
| [28] | Trp-P-2 acetate | 72254-58-1 | Wako | 97 | 25 | |||||
| [29] | MeAαC acetate | 68006-83-7a | Wako | 99 | 14 | |||||
| [30] | IQ | 76180-96-6 | Wako | >99 | 7 | |||||
| [31] | MeIQx | 77500-04-0 | Wako | 99 | 1 | |||||
| Aromatic nitro compounds | ||||||||||
| [32] | Chloramphenicol | 56-75-7 | Sigma | 98 | 18 | |||||
| [33] | Metronidazole | 443-48-1 | Tokyo Kasei | 100 | 18 | |||||
| [34] | 2,4-Dinitrotoluene | 121-14-2 | Wako | 99 | 23 | |||||
| [35] | 2-Nitrofluorene | 607-57-8 | Tokyo Kasei | 99 | 11 | |||||
| [36] | 1-Nitronaphthalene | 86-57-7 | Kanto Chemical | 98 | 11 | |||||
| [37] | Azathioprine | 446-86-6 | Sigma | 98 | 3 | |||||
| [38] | AF-2 | 3688-53-7 | Wako | 99 | 23 | |||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | 56-57-5 | Tokyo Kasei | 98 | 5 | |||||
| [40] | 1-Nitropyrene | 5522-43-0 | Aldrich | 99 | 3 | |||||
| Azo dyes | ||||||||||
| [41] | Azobenzene | 103-33-3 | Tokyo Kasei | 100 | 2 | |||||
| [42] | 4-Aminoazobenzene | 60-09-3 | Tokyo Kasei | 100 | 2 | |||||
| [43] | o-Aminoazotoluene | 97-56-3 | Tokyo Kasei | >95 | 13 | |||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 60-11-7 | Tokyo Kasei | GRb | 2 | |||||
| [45] | Congo red (Direct Red 28) | 573-58-0 | Wako | GR | 9 | |||||
| [46] | Trypan blue | 72-57-1 | Aldrich | 96 | 21 | |||||
| [46′] | Trypan blue (crude) | 72-57-1 | Nacalai | GR | 21 | |||||
| Nitrosamines | ||||||||||
| [47] | N-Nitrosodimethylamine | 62-75-9 | Tokyo Kasei | 100 | 4 | |||||
| [48] | N-Nitrosodiethylamine | 55-18-5 | Tokyo Kasei | >99 | 4 | |||||
| [49] | N-Nitroso-di-n-propylamine | 621-64-7 | Tokyo Kasei | 100 | 4 | |||||
| [50] | N-Nitroso-di-n-butylamine | 924-16-3 | Tokyo Kasei | 100 | 4 | |||||
| [51] | N-Nitrosomorpholine | 59-89-2 | Tokyo Kasei | 100 | 4 | |||||
| Vinyl compounds | ||||||||||
| [52] | Acrylonitrile | 107-13-1 | Tokyo Kasei | >99 | 15 | |||||
| [53] | Acrylamide | 79-06-1 | Nacalai | 100 | 20 | |||||
| [54] | Styrene | 100-42-5 | Tokyo Kasei | >99 | 9 | |||||
| [55] | Safrole | 94-59-7 | Tokyo Kasei | >96 | 19 | |||||
| Miscellaneous nitrogen compounds | ||||||||||
| [56] | Hydrazine 2HCl | 5341-61-7 | Tokyo Kasei | >98 | 6 | |||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 306-37-6 | Tokyo Kasei | >98 | 6 | |||||
| [58] | Dacarbazine | 4342-03-4 | Sigma | 98 | 12 | |||||
| [59] | Sodium nitrite | 7632-00-0 | Wako | 99 | 16 | |||||
| [60] | Sodium nitrate | 7631-99-4 | Wako | 99 | 16 | |||||
Free form.
GR; manufacturers' guaranteed reagent. The purity of the crude trypan blue test article was roughly estimated 50%, although not specifically determined.
Chemical mutagens assayed in this study
| Chemical no. . | Chemical name . | CAS no. . | Source . | Purity (%) . | Assay lab no. . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polycyclic hydrocarbons | ||||||||||
| [1] | Phenanthrene | 85-01-8 | Tokyo Kasei | >98 | 7 | |||||
| [2] | Pyrene | 129-00-0 | Tokyo Kasei | >97 | 17 | |||||
| [3] | Chrysene | 218-01-9 | Tokyo Kasei | 99 | 10 | |||||
| [4] | Benz[a]anthracene | 56-55-3 | Tokyo Kasei | >99 | 25 | |||||
| [5] | Dibenz[a,c]anthracene | 215-58-7 | Tokyo Kasei | >99 | 15 | |||||
| [6] | Dibenz[a,h]anthracene | 53-70-3 | Tokyo Kasei | >95 | 17 | |||||
| [7] | Benzo[a]pyrene | 50-32-8 | Tokyo Kasei | >95 | 4 | |||||
| [8] | 7,12-Dimethylbenz[a]anthracene | 57-97-6 | Nacalai | 98 | 20 | |||||
| [9] | 3-Methylcholanthrene | 56-49-5 | Aldrich | 98 | 8 | |||||
| Heterocyclic hydrocarbons | ||||||||||
| [10] | Quinoline | 91-22-5 | Sigma | 96 | 24 | |||||
| [11] | 1,7-Phenanthroline | 230-46-6 | Aldrich | 100 | 14 | |||||
| [12] | 4-Methylquinoline (Lepidine) | 491-35-0 | Sigma | 99 | 24 | |||||
| [13] | 7,9-Dimethylbenz[c]acridine | 963-89-3 | Aldrich | 92 | 10 | |||||
| [14] | Aflatoxin B1 | 1162-65-8 | Wako | >97 | 4 | |||||
| [15] | Quercetin | 117-39-5 | Tokyo Kasei | 100 | 4 | |||||
| Aromatic amines | ||||||||||
| [16] | 2,4-Diaminotoluene | 95-80-7 | Tokyo Kasei | 98 | 5 | |||||
| [17] | 3,3′-Dimethylbenzidine (o-Tolidine) | 119-93-7 | Wako | >97 | 21 | |||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 91-94-1 | Sigma | >99 | 8 | |||||
| [19] | 4,4′-Methylenedianiline | 101-77-9 | Tokyo Kasei | 100 | 12 | |||||
| [20] | Auramine (Basic Yellow 2) | 2465-27-2 | Tokyo Kasei | 81 | 22 | |||||
| [21] | 1-Naphthylamine HCl | 552-46-5 | Tokyo Kasei | >97 | 22 | |||||
| [22] | 2-Aminofluorene | 153-78-6 | Wako | >97 | 13 | |||||
| [23] | 2-Acetylaminofluorene | 53-96-3 | Tokyo Kasei | >98 | 4 | |||||
| [24] | 2-Aminoanthracene | 613-13-8 | Wako | 95 | 4 | |||||
| [25] | 6-Aminochrysene | 2642-98-0 | Aldrich | 97 | 4 | |||||
| [26] | 1-Aminopyrene | 1606-67-3 | Tokyo Kasei | >95 | 19 | |||||
| Heterocyclic amines | ||||||||||
| [27] | PhIP HCl | 105650-23-5 | Wako | 99 | 1 | |||||
| [28] | Trp-P-2 acetate | 72254-58-1 | Wako | 97 | 25 | |||||
| [29] | MeAαC acetate | 68006-83-7a | Wako | 99 | 14 | |||||
| [30] | IQ | 76180-96-6 | Wako | >99 | 7 | |||||
| [31] | MeIQx | 77500-04-0 | Wako | 99 | 1 | |||||
| Aromatic nitro compounds | ||||||||||
| [32] | Chloramphenicol | 56-75-7 | Sigma | 98 | 18 | |||||
| [33] | Metronidazole | 443-48-1 | Tokyo Kasei | 100 | 18 | |||||
| [34] | 2,4-Dinitrotoluene | 121-14-2 | Wako | 99 | 23 | |||||
| [35] | 2-Nitrofluorene | 607-57-8 | Tokyo Kasei | 99 | 11 | |||||
| [36] | 1-Nitronaphthalene | 86-57-7 | Kanto Chemical | 98 | 11 | |||||
| [37] | Azathioprine | 446-86-6 | Sigma | 98 | 3 | |||||
| [38] | AF-2 | 3688-53-7 | Wako | 99 | 23 | |||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | 56-57-5 | Tokyo Kasei | 98 | 5 | |||||
| [40] | 1-Nitropyrene | 5522-43-0 | Aldrich | 99 | 3 | |||||
| Azo dyes | ||||||||||
| [41] | Azobenzene | 103-33-3 | Tokyo Kasei | 100 | 2 | |||||
| [42] | 4-Aminoazobenzene | 60-09-3 | Tokyo Kasei | 100 | 2 | |||||
| [43] | o-Aminoazotoluene | 97-56-3 | Tokyo Kasei | >95 | 13 | |||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 60-11-7 | Tokyo Kasei | GRb | 2 | |||||
| [45] | Congo red (Direct Red 28) | 573-58-0 | Wako | GR | 9 | |||||
| [46] | Trypan blue | 72-57-1 | Aldrich | 96 | 21 | |||||
| [46′] | Trypan blue (crude) | 72-57-1 | Nacalai | GR | 21 | |||||
| Nitrosamines | ||||||||||
| [47] | N-Nitrosodimethylamine | 62-75-9 | Tokyo Kasei | 100 | 4 | |||||
| [48] | N-Nitrosodiethylamine | 55-18-5 | Tokyo Kasei | >99 | 4 | |||||
| [49] | N-Nitroso-di-n-propylamine | 621-64-7 | Tokyo Kasei | 100 | 4 | |||||
| [50] | N-Nitroso-di-n-butylamine | 924-16-3 | Tokyo Kasei | 100 | 4 | |||||
| [51] | N-Nitrosomorpholine | 59-89-2 | Tokyo Kasei | 100 | 4 | |||||
| Vinyl compounds | ||||||||||
| [52] | Acrylonitrile | 107-13-1 | Tokyo Kasei | >99 | 15 | |||||
| [53] | Acrylamide | 79-06-1 | Nacalai | 100 | 20 | |||||
| [54] | Styrene | 100-42-5 | Tokyo Kasei | >99 | 9 | |||||
| [55] | Safrole | 94-59-7 | Tokyo Kasei | >96 | 19 | |||||
| Miscellaneous nitrogen compounds | ||||||||||
| [56] | Hydrazine 2HCl | 5341-61-7 | Tokyo Kasei | >98 | 6 | |||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 306-37-6 | Tokyo Kasei | >98 | 6 | |||||
| [58] | Dacarbazine | 4342-03-4 | Sigma | 98 | 12 | |||||
| [59] | Sodium nitrite | 7632-00-0 | Wako | 99 | 16 | |||||
| [60] | Sodium nitrate | 7631-99-4 | Wako | 99 | 16 | |||||
| Chemical no. . | Chemical name . | CAS no. . | Source . | Purity (%) . | Assay lab no. . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polycyclic hydrocarbons | ||||||||||
| [1] | Phenanthrene | 85-01-8 | Tokyo Kasei | >98 | 7 | |||||
| [2] | Pyrene | 129-00-0 | Tokyo Kasei | >97 | 17 | |||||
| [3] | Chrysene | 218-01-9 | Tokyo Kasei | 99 | 10 | |||||
| [4] | Benz[a]anthracene | 56-55-3 | Tokyo Kasei | >99 | 25 | |||||
| [5] | Dibenz[a,c]anthracene | 215-58-7 | Tokyo Kasei | >99 | 15 | |||||
| [6] | Dibenz[a,h]anthracene | 53-70-3 | Tokyo Kasei | >95 | 17 | |||||
| [7] | Benzo[a]pyrene | 50-32-8 | Tokyo Kasei | >95 | 4 | |||||
| [8] | 7,12-Dimethylbenz[a]anthracene | 57-97-6 | Nacalai | 98 | 20 | |||||
| [9] | 3-Methylcholanthrene | 56-49-5 | Aldrich | 98 | 8 | |||||
| Heterocyclic hydrocarbons | ||||||||||
| [10] | Quinoline | 91-22-5 | Sigma | 96 | 24 | |||||
| [11] | 1,7-Phenanthroline | 230-46-6 | Aldrich | 100 | 14 | |||||
| [12] | 4-Methylquinoline (Lepidine) | 491-35-0 | Sigma | 99 | 24 | |||||
| [13] | 7,9-Dimethylbenz[c]acridine | 963-89-3 | Aldrich | 92 | 10 | |||||
| [14] | Aflatoxin B1 | 1162-65-8 | Wako | >97 | 4 | |||||
| [15] | Quercetin | 117-39-5 | Tokyo Kasei | 100 | 4 | |||||
| Aromatic amines | ||||||||||
| [16] | 2,4-Diaminotoluene | 95-80-7 | Tokyo Kasei | 98 | 5 | |||||
| [17] | 3,3′-Dimethylbenzidine (o-Tolidine) | 119-93-7 | Wako | >97 | 21 | |||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 91-94-1 | Sigma | >99 | 8 | |||||
| [19] | 4,4′-Methylenedianiline | 101-77-9 | Tokyo Kasei | 100 | 12 | |||||
| [20] | Auramine (Basic Yellow 2) | 2465-27-2 | Tokyo Kasei | 81 | 22 | |||||
| [21] | 1-Naphthylamine HCl | 552-46-5 | Tokyo Kasei | >97 | 22 | |||||
| [22] | 2-Aminofluorene | 153-78-6 | Wako | >97 | 13 | |||||
| [23] | 2-Acetylaminofluorene | 53-96-3 | Tokyo Kasei | >98 | 4 | |||||
| [24] | 2-Aminoanthracene | 613-13-8 | Wako | 95 | 4 | |||||
| [25] | 6-Aminochrysene | 2642-98-0 | Aldrich | 97 | 4 | |||||
| [26] | 1-Aminopyrene | 1606-67-3 | Tokyo Kasei | >95 | 19 | |||||
| Heterocyclic amines | ||||||||||
| [27] | PhIP HCl | 105650-23-5 | Wako | 99 | 1 | |||||
| [28] | Trp-P-2 acetate | 72254-58-1 | Wako | 97 | 25 | |||||
| [29] | MeAαC acetate | 68006-83-7a | Wako | 99 | 14 | |||||
| [30] | IQ | 76180-96-6 | Wako | >99 | 7 | |||||
| [31] | MeIQx | 77500-04-0 | Wako | 99 | 1 | |||||
| Aromatic nitro compounds | ||||||||||
| [32] | Chloramphenicol | 56-75-7 | Sigma | 98 | 18 | |||||
| [33] | Metronidazole | 443-48-1 | Tokyo Kasei | 100 | 18 | |||||
| [34] | 2,4-Dinitrotoluene | 121-14-2 | Wako | 99 | 23 | |||||
| [35] | 2-Nitrofluorene | 607-57-8 | Tokyo Kasei | 99 | 11 | |||||
| [36] | 1-Nitronaphthalene | 86-57-7 | Kanto Chemical | 98 | 11 | |||||
| [37] | Azathioprine | 446-86-6 | Sigma | 98 | 3 | |||||
| [38] | AF-2 | 3688-53-7 | Wako | 99 | 23 | |||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | 56-57-5 | Tokyo Kasei | 98 | 5 | |||||
| [40] | 1-Nitropyrene | 5522-43-0 | Aldrich | 99 | 3 | |||||
| Azo dyes | ||||||||||
| [41] | Azobenzene | 103-33-3 | Tokyo Kasei | 100 | 2 | |||||
| [42] | 4-Aminoazobenzene | 60-09-3 | Tokyo Kasei | 100 | 2 | |||||
| [43] | o-Aminoazotoluene | 97-56-3 | Tokyo Kasei | >95 | 13 | |||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 60-11-7 | Tokyo Kasei | GRb | 2 | |||||
| [45] | Congo red (Direct Red 28) | 573-58-0 | Wako | GR | 9 | |||||
| [46] | Trypan blue | 72-57-1 | Aldrich | 96 | 21 | |||||
| [46′] | Trypan blue (crude) | 72-57-1 | Nacalai | GR | 21 | |||||
| Nitrosamines | ||||||||||
| [47] | N-Nitrosodimethylamine | 62-75-9 | Tokyo Kasei | 100 | 4 | |||||
| [48] | N-Nitrosodiethylamine | 55-18-5 | Tokyo Kasei | >99 | 4 | |||||
| [49] | N-Nitroso-di-n-propylamine | 621-64-7 | Tokyo Kasei | 100 | 4 | |||||
| [50] | N-Nitroso-di-n-butylamine | 924-16-3 | Tokyo Kasei | 100 | 4 | |||||
| [51] | N-Nitrosomorpholine | 59-89-2 | Tokyo Kasei | 100 | 4 | |||||
| Vinyl compounds | ||||||||||
| [52] | Acrylonitrile | 107-13-1 | Tokyo Kasei | >99 | 15 | |||||
| [53] | Acrylamide | 79-06-1 | Nacalai | 100 | 20 | |||||
| [54] | Styrene | 100-42-5 | Tokyo Kasei | >99 | 9 | |||||
| [55] | Safrole | 94-59-7 | Tokyo Kasei | >96 | 19 | |||||
| Miscellaneous nitrogen compounds | ||||||||||
| [56] | Hydrazine 2HCl | 5341-61-7 | Tokyo Kasei | >98 | 6 | |||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 306-37-6 | Tokyo Kasei | >98 | 6 | |||||
| [58] | Dacarbazine | 4342-03-4 | Sigma | 98 | 12 | |||||
| [59] | Sodium nitrite | 7632-00-0 | Wako | 99 | 16 | |||||
| [60] | Sodium nitrate | 7631-99-4 | Wako | 99 | 16 | |||||
Free form.
GR; manufacturers' guaranteed reagent. The purity of the crude trypan blue test article was roughly estimated 50%, although not specifically determined.
Bacterial strains
The test strains used for the Ames test were TA100 (hisG46/rfa/ΔuvrB/pKM101), TA98 (hisD3052/rfa/ΔuvrB/pKM101) or YG7108 (hisG46/rfa/ΔuvrB/ΔadaST/ΔogtST). The TA100 and TA98 strains, developed by Dr B.N.Ames of the University of California, Berkeley, detect base-pair substitution mutations and frameshift mutations, respectively (1). The YG7108 strain, developed by Drs T.Nohmi and M.Yamada of the National Institute of Health Sciences, is highly sensitive to alkylating agents (32). The characteristics of these test strains, including their susceptibility to mutagens, were confirmed prior to use in the assays in each laboratory.
Preparation of liver S9 fractions
Pooled S9 and HLS-014 were prepared according to the S9 preparation procedures with a modified re-centrifugation step (27,29) at the Biomedical Research Institute of the Human and Animal Bridging Research Organization (HAB) in Chiba, Japan, using frozen human livers. Pooled S9 was the S9 fraction prepared from 15 different donor liver tissues (most of the donors were Caucasians), while HLS-014 was the S9 fraction prepared from a human liver that showed the highest ability to induce the mutagenicity of three procarcinogens among the 18 donor liver tissues stocked in the Biomedical Research Institute (28). These samples were obtained from non-transplantable liver donors that, because of certain medical reason(s) such as immunological incongruence, could not be used. Liver samples were legally procured from the NDRI (National Disease Research Interchange) in Philadelphia, USA, with permission to use for research purposes only, based on the international partnership between the NDRI and HAB in Japan. Rat S9, prepared from male Sprague–Dawley rats intraperitoneally treated with PB at doses of 30 mg/kg/day (96 h before killing) and 60 mg/kg/day (24, 48 and 72 h before killing) and with BF at a dose of 80 mg/kg/day (48 h before killing), was purchased from Oriental Yeast Co., Tokyo, Japan. Analytical data for the two human and rat liver S9 fractions are shown in Table III.
Contents and enzyme activities of P450s in human and rat liver S9 fractions
| S9 fraction . | Total P450 contenta (pmol/mg protein) . | Enzyme activityb (pmol/min/mg protein) . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Ethoxyresorufin O-deethylationc . | Chlorzoxazone 6-hydroxylationd . | Testosterone 6β-hydroxylatione . | ||
| Human S9 (pooled) | 44 (1.0)f | 90 (1.0) | 487 (1.0) | 824 (1.0) | ||
| Human S9 (HLS-014) | 119 (2.7) | 429 (4.8) | 535 (1.1) | 4326 (5.3) | ||
| PB/BF-induced rat S9 | 715 (16) | 4670 (52) | 1295 (2.7) | 2733 (3.3) | ||
| S9 fraction . | Total P450 contenta (pmol/mg protein) . | Enzyme activityb (pmol/min/mg protein) . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Ethoxyresorufin O-deethylationc . | Chlorzoxazone 6-hydroxylationd . | Testosterone 6β-hydroxylatione . | ||
| Human S9 (pooled) | 44 (1.0)f | 90 (1.0) | 487 (1.0) | 824 (1.0) | ||
| Human S9 (HLS-014) | 119 (2.7) | 429 (4.8) | 535 (1.1) | 4326 (5.3) | ||
| PB/BF-induced rat S9 | 715 (16) | 4670 (52) | 1295 (2.7) | 2733 (3.3) | ||
Measured by the method of Omura and Sato (33).
Determined according to the method of Ikeda et al. (34) using HPLC procedures.
Each value represents a specific activity for CYP1A1/2.
Each value represents a specific activity for CYP2E1.
Each value represents a specific activity for CYP3A.
Figures in parentheses indicate the ratio relative to human S9 (pooled).
Contents and enzyme activities of P450s in human and rat liver S9 fractions
| S9 fraction . | Total P450 contenta (pmol/mg protein) . | Enzyme activityb (pmol/min/mg protein) . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Ethoxyresorufin O-deethylationc . | Chlorzoxazone 6-hydroxylationd . | Testosterone 6β-hydroxylatione . | ||
| Human S9 (pooled) | 44 (1.0)f | 90 (1.0) | 487 (1.0) | 824 (1.0) | ||
| Human S9 (HLS-014) | 119 (2.7) | 429 (4.8) | 535 (1.1) | 4326 (5.3) | ||
| PB/BF-induced rat S9 | 715 (16) | 4670 (52) | 1295 (2.7) | 2733 (3.3) | ||
| S9 fraction . | Total P450 contenta (pmol/mg protein) . | Enzyme activityb (pmol/min/mg protein) . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Ethoxyresorufin O-deethylationc . | Chlorzoxazone 6-hydroxylationd . | Testosterone 6β-hydroxylatione . | ||
| Human S9 (pooled) | 44 (1.0)f | 90 (1.0) | 487 (1.0) | 824 (1.0) | ||
| Human S9 (HLS-014) | 119 (2.7) | 429 (4.8) | 535 (1.1) | 4326 (5.3) | ||
| PB/BF-induced rat S9 | 715 (16) | 4670 (52) | 1295 (2.7) | 2733 (3.3) | ||
Measured by the method of Omura and Sato (33).
Determined according to the method of Ikeda et al. (34) using HPLC procedures.
Each value represents a specific activity for CYP1A1/2.
Each value represents a specific activity for CYP2E1.
Each value represents a specific activity for CYP3A.
Figures in parentheses indicate the ratio relative to human S9 (pooled).
Mutagenicity assay
The Ames test (the pre-incubation method at 37°C for 20 min) was conducted to examine the mutagenicity of the chemicals (1,26,28,29). Briefly, a mixture containing each test compound in 0.1 ml of dimethyl sulfoxide (DMSO), 0.1 ml of the test strain cell culture in the early stationary phase and 0.5 ml of S9 mix was incubated at 37°C for 20 min in each test tube with a shaking frequency of 120 strokes per minute. After incubation, 2 ml of 0.05 mM l-histidine/0.05 mM biotin molten top agar were added to each test tube, mixed and poured onto the surface of minimal glucose agar medium. In the case of acrylonitrile, the test chemical was removed by discarding the supernatant after centrifugation prior to combination with the top agar. The plate was incubated for ∼48 h at 37°C and the number of revertant colonies was counted. The Ames test using the three different S9 fractions for all the test compounds and without an S9 mix (phosphate buffer in place of the S9 mix) for several compounds was performed on the same occasion in each laboratory.
The S9 mix (0.5 ml) contained 0.05 ml of the S9 fraction and 0.45 ml of a cofactor solution (Cofactor I, Oriental Yeast Co., Ltd). The S9 mix composed of 8 mM MgCl2, 33 mM KCl, 5 mM glucose-6-phosphate, 4 mM NADPH, 4 mM NADH and 100 mM sodium phosphate (pH 7.4). The protein amount of each S9 fraction that was used was 1 mg/plate.
To validate the assay system employed in this collaborative study, a positive control (benzo[a]pyrene at 27 µg/plate for TA100) was used and the sterility of each S9 fraction was checked. The dose-finding and main assays were conducted using one plate and two plates, respectively. DMSO was used to dissolve the test and positive control articles and was also used as a negative (solvent) control.
Determination of total P450 content and cytochrome P450 activities
The total P450 content of the S9 fractions was measured using the method of Omura and Sato (33). The cytochrome P450 activities of the S9 fractions were determined according to the method of Ikeda et al. (34), using HPLC procedures.
Evaluation of mutagenicity
Mutagenicity was evaluated according to the so-called ‘2-fold’ rule (2). Thus, chemicals were judged to be mutagenic if the following criteria were satisfied: (1) the maximum number of revertants was 2-fold or more relative to the negative control, (2) a dose-dependent increase in the number of revertants was observed and (3) the dose-finding and main assays produced reproducible results. If no increase in the number of revertants was observed, the chemicals were judged to have a negative result. If the ‘mutagenic’ and ‘negative’ criteria were not met, then the chemicals were judged to have exhibited an equivocal response. Mutagenic activity (equal to the number of induced revertants/µg/plate) was calculated as a measure of mutagenicity (Table IV). Thus, in the case of mutagenic chemicals, mutagenic activity was expressed as the greatest value of revertants per µg of test compound, as calculated from each point on the dose–response curves, presented in Figure 1. If the chemicals showed an equivocal response and no mutagenicity, this was marked by a ‘±’ and ‘0’, respectively, in this text.
Individual data on the mutagenicity of 58 compounds tested in the presence of the HLS-014, pooled S9 and rat S9 fractions at an equivalent amount of S9 protein (1 mg/plate). Mutagenicity was assayed using the Ames test (the pre-incubation method at 37°C for 20 min) with S.typhimurium TA100, TA98 or YG7108. Assays were conducted using two plates, and the mean values were plotted. The symbols open triangle (▵), closed square (▪), open circle (○), and closed circle (•) used in the dose–response curves indicate treatments without S9 mix, with rat S9, with HLS-014 and with pooled S9, respectively.
Summary data of the Salmonella/human S9 mutagenicity test
| Chemical no. . | Chemical name . | IARC groupa . | Bacterial strain . | Mutagenic activity (induced revertants/µg/plate) . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | . | -S9 mix . | Induced rat S9 . | Human S9 (HLS-014) . | Human S9 (pooled) . | |||||||
| Polycyclic hydrocarbons | ||||||||||||||
| [1] | Phenanthrene | 3 | TA100 | 0 | 0 | 0 | 0 | |||||||
| [2] | Pyrene | 3 | TA100 | NTb | 0 | 0 | 0 | |||||||
| [3] | Chrysene | 3 | TA100 | NT | 110 | ±c | 0 | |||||||
| [4] | Benz[a]anthracene | 2A | TA100 | NT | 23 | 0 | 0 | |||||||
| [5] | Dibenz[a,c]anthracene | 3 | TA100 | NT | 530 | 0 | 0 | |||||||
| [6] | Dibenz[a,h]anthracene | 2A | TA100 | NT | 2.6 | 0 | 0 | |||||||
| [7]d | Benzo[a]pyrene | 2A | TA100 | NT | 322 | 226 | ± | |||||||
| [8] | 7,12-Dimethylbenz[a]anthracene | nc | TA100 | 11 | 190 | 480 | 51 | |||||||
| [9] | 3-Methylcholanthrene | nc | TA100 | NT | 260 | 200 | ± | |||||||
| Heterocyclic hydrocarbons | ||||||||||||||
| [10] | Quinoline | nc | TA100 | 0 | 3.0 | 1.6 | 0.83 | |||||||
| [11] | 1,7-Phenanthroline | nc | TA100 | NT | 12 | 0 | 0 | |||||||
| [12] | 4-Methylquinoline | nc | TA100 | 0 | 14 | 15 | 3.0 | |||||||
| [13] | 7,9-Dimethylbenz[c]acridine | nc | TA100 | NT | 150 | 630 | ± | |||||||
| [14] | Aflatoxin B1 | 1 | TA100 | 97 | 67000 | 4000 | 810 | |||||||
| [15] | Quercetin | 3 | TA100 | NT | 36 | 40 | 37 | |||||||
| Aromatic amines | ||||||||||||||
| [16] | 2,4-Diaminotoluene | 2B | TA100 | NT | 0.044 | ± | 0 | |||||||
| [17] | o-Tolidine | 2B | TA98 | 0 | 0.3 | 0 | 0 | |||||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 2B | TA98 | NT | 210 | 160 | 27 | |||||||
| [19] | 4,4′-Methylenedianiline | 2B | TA100 | 0 | 5.1 | 4.6 | 0.11 | |||||||
| [20] | Auramine (Basic Yellow 2) | 2B | TA100 | 0 | 0 | 0 | 0 | |||||||
| [21] | 1-Naphthylamine HCl | 3 | TA100 | 0 | ± | 10 | 1.8 | |||||||
| [22] | 2-Aminofluorene | nc | TA100 | ± | 200 | 5000 | 200 | |||||||
| [23] | 2-Acetylaminofluorene | nc | TA100 | 0 | 59 | 390 | 29 | |||||||
| [24] | 2-Aminoanthracene | nc | TA100 | 0 | 1500 | 24000 | 22000 | |||||||
| [25] | 6-Aminochrysene | nc | TA100 | 21 | 4000 | 5000 | 500 | |||||||
| [26] | 1-Aminopyrene | nc | TA100 | NT | 390 | 190 | ± | |||||||
| Heterocyclic amines | ||||||||||||||
| [27] | PhIP HCl | 2B | TA100 | NT | 260 | 35 | 0.32 | |||||||
| [28] | Trp-P-2 acetate | 2B | TA100 | NT | 1400 | ± | ± | |||||||
| [29] | MeAαC acetate | 2B | TA100 | NT | 260 | 56 | 5.6 | |||||||
| [30] | IQ | 2A | TA100 | NT | 18000 | 6000 | 340 | |||||||
| [31] | MeIQx | 2B | TA100 | NT | 4000 | 2200 | 84 | |||||||
| Aromatic nitro compounds | ||||||||||||||
| [32] | Chloramphenicol | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [33] | Metronidazole | 2B | TA100 | 5.7 | 6.0 | 4.8 | 5.0 | |||||||
| [34] | 2,4-Dinitrotoluene | 2B | TA100 | 0.58 | 0.46 | 0.33 | 0.28 | |||||||
| [35] | 2-Nitrofluorene | 2B | TA100 | 560 | 430 | 460 | 800 | |||||||
| [36] | 1-Nitronaphthalene | 3 | TA100 | 34 | 19 | 58 | 80 | |||||||
| [37] | Azathioprine | 1 | TA100 | 14 | 4.3 | 3.1 | 2.2 | |||||||
| [38] | AF-2 | 2B | TA100 | 79000 | 15000 | 31000 | 41000 | |||||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | nc | TA100 | 8400 | 380 | 3200 | 3700 | |||||||
| [40] | 1-Nitropyrene | 2B | TA100 | 1000 | 280 | 1400 | 950 | |||||||
| Azo dyes | ||||||||||||||
| [41] | Azobenzene | 3 | TA100 | NT | 5.1 | 2.9 | 0 | |||||||
| [42] | 4-Aminoazobenzene | 2B | TA100 | NT | 4.6 | 1.4 | 0 | |||||||
| [43] | o-Aminoazotoluene | 2B | TA100 | 0 | 69 | 25 | ± | |||||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [45] | Congo red (Direct Red 28) | nc | TA100 | NT | 0 | 0 | 0 | |||||||
| [46a] | Trypan blue | 2B | TA98 | NT | 0 | 0 | 0 | |||||||
| [46b] | Trypan blue (crude) | 2B | TA98 | 0 | 0.14 | 1.0 | 0.49 | |||||||
| Nitrosamines | ||||||||||||||
| [47]d | N-Nitrosodimethylamine | 2A | YG7108 | NT | 8.0 | 66 | 63 | |||||||
| [48] | N-Nitrosodiethylamine | 2A | YG7108 | ± | 4.3 | 11 | 6.4 | |||||||
| [49] | N-Nitroso-di-n-propylamine | 2B | YG7108 | NT | 25 | 2.5 | 1.0 | |||||||
| [50] | N-Nitroso-di-n-butylamine | 2B | YG7108 | NT | 52 | 0.52 | ± | |||||||
| [51] | N-Nitrosomorpholine | 2B | YG7108 | NT | 0.56 | 0.63 | 0.33 | |||||||
| Vinyl compounds | ||||||||||||||
| [52] | Acrylonitrile | 2B | TA100 | NT | 0.0093 | 0.0082 | 0.0088 | |||||||
| [53] | Acrylamide | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [54] | Styrene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [55] | Safrole | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| Miscellaneous nitrogen compounds | ||||||||||||||
| [56] | Hydrazine 2HCl | 2B | TA100 | 0 | 0.16 | 0.18 | 0.10 | |||||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 2A | TA100 | ± | ± | ± | ± | |||||||
| [58] | Dacarbazine | 2B | TA100 | 1.0 | 0.55 | 0.93 | 1.1 | |||||||
| [59] | Sodium nitrite | nc | TA100 | 0.065 | 0.051 | 0.052 | 0.056 | |||||||
| [60] | Sodium nitrate | nc | TA100 | 0 | 0 | 0 | 0 | |||||||
| Chemical no. . | Chemical name . | IARC groupa . | Bacterial strain . | Mutagenic activity (induced revertants/µg/plate) . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | . | -S9 mix . | Induced rat S9 . | Human S9 (HLS-014) . | Human S9 (pooled) . | |||||||
| Polycyclic hydrocarbons | ||||||||||||||
| [1] | Phenanthrene | 3 | TA100 | 0 | 0 | 0 | 0 | |||||||
| [2] | Pyrene | 3 | TA100 | NTb | 0 | 0 | 0 | |||||||
| [3] | Chrysene | 3 | TA100 | NT | 110 | ±c | 0 | |||||||
| [4] | Benz[a]anthracene | 2A | TA100 | NT | 23 | 0 | 0 | |||||||
| [5] | Dibenz[a,c]anthracene | 3 | TA100 | NT | 530 | 0 | 0 | |||||||
| [6] | Dibenz[a,h]anthracene | 2A | TA100 | NT | 2.6 | 0 | 0 | |||||||
| [7]d | Benzo[a]pyrene | 2A | TA100 | NT | 322 | 226 | ± | |||||||
| [8] | 7,12-Dimethylbenz[a]anthracene | nc | TA100 | 11 | 190 | 480 | 51 | |||||||
| [9] | 3-Methylcholanthrene | nc | TA100 | NT | 260 | 200 | ± | |||||||
| Heterocyclic hydrocarbons | ||||||||||||||
| [10] | Quinoline | nc | TA100 | 0 | 3.0 | 1.6 | 0.83 | |||||||
| [11] | 1,7-Phenanthroline | nc | TA100 | NT | 12 | 0 | 0 | |||||||
| [12] | 4-Methylquinoline | nc | TA100 | 0 | 14 | 15 | 3.0 | |||||||
| [13] | 7,9-Dimethylbenz[c]acridine | nc | TA100 | NT | 150 | 630 | ± | |||||||
| [14] | Aflatoxin B1 | 1 | TA100 | 97 | 67000 | 4000 | 810 | |||||||
| [15] | Quercetin | 3 | TA100 | NT | 36 | 40 | 37 | |||||||
| Aromatic amines | ||||||||||||||
| [16] | 2,4-Diaminotoluene | 2B | TA100 | NT | 0.044 | ± | 0 | |||||||
| [17] | o-Tolidine | 2B | TA98 | 0 | 0.3 | 0 | 0 | |||||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 2B | TA98 | NT | 210 | 160 | 27 | |||||||
| [19] | 4,4′-Methylenedianiline | 2B | TA100 | 0 | 5.1 | 4.6 | 0.11 | |||||||
| [20] | Auramine (Basic Yellow 2) | 2B | TA100 | 0 | 0 | 0 | 0 | |||||||
| [21] | 1-Naphthylamine HCl | 3 | TA100 | 0 | ± | 10 | 1.8 | |||||||
| [22] | 2-Aminofluorene | nc | TA100 | ± | 200 | 5000 | 200 | |||||||
| [23] | 2-Acetylaminofluorene | nc | TA100 | 0 | 59 | 390 | 29 | |||||||
| [24] | 2-Aminoanthracene | nc | TA100 | 0 | 1500 | 24000 | 22000 | |||||||
| [25] | 6-Aminochrysene | nc | TA100 | 21 | 4000 | 5000 | 500 | |||||||
| [26] | 1-Aminopyrene | nc | TA100 | NT | 390 | 190 | ± | |||||||
| Heterocyclic amines | ||||||||||||||
| [27] | PhIP HCl | 2B | TA100 | NT | 260 | 35 | 0.32 | |||||||
| [28] | Trp-P-2 acetate | 2B | TA100 | NT | 1400 | ± | ± | |||||||
| [29] | MeAαC acetate | 2B | TA100 | NT | 260 | 56 | 5.6 | |||||||
| [30] | IQ | 2A | TA100 | NT | 18000 | 6000 | 340 | |||||||
| [31] | MeIQx | 2B | TA100 | NT | 4000 | 2200 | 84 | |||||||
| Aromatic nitro compounds | ||||||||||||||
| [32] | Chloramphenicol | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [33] | Metronidazole | 2B | TA100 | 5.7 | 6.0 | 4.8 | 5.0 | |||||||
| [34] | 2,4-Dinitrotoluene | 2B | TA100 | 0.58 | 0.46 | 0.33 | 0.28 | |||||||
| [35] | 2-Nitrofluorene | 2B | TA100 | 560 | 430 | 460 | 800 | |||||||
| [36] | 1-Nitronaphthalene | 3 | TA100 | 34 | 19 | 58 | 80 | |||||||
| [37] | Azathioprine | 1 | TA100 | 14 | 4.3 | 3.1 | 2.2 | |||||||
| [38] | AF-2 | 2B | TA100 | 79000 | 15000 | 31000 | 41000 | |||||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | nc | TA100 | 8400 | 380 | 3200 | 3700 | |||||||
| [40] | 1-Nitropyrene | 2B | TA100 | 1000 | 280 | 1400 | 950 | |||||||
| Azo dyes | ||||||||||||||
| [41] | Azobenzene | 3 | TA100 | NT | 5.1 | 2.9 | 0 | |||||||
| [42] | 4-Aminoazobenzene | 2B | TA100 | NT | 4.6 | 1.4 | 0 | |||||||
| [43] | o-Aminoazotoluene | 2B | TA100 | 0 | 69 | 25 | ± | |||||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [45] | Congo red (Direct Red 28) | nc | TA100 | NT | 0 | 0 | 0 | |||||||
| [46a] | Trypan blue | 2B | TA98 | NT | 0 | 0 | 0 | |||||||
| [46b] | Trypan blue (crude) | 2B | TA98 | 0 | 0.14 | 1.0 | 0.49 | |||||||
| Nitrosamines | ||||||||||||||
| [47]d | N-Nitrosodimethylamine | 2A | YG7108 | NT | 8.0 | 66 | 63 | |||||||
| [48] | N-Nitrosodiethylamine | 2A | YG7108 | ± | 4.3 | 11 | 6.4 | |||||||
| [49] | N-Nitroso-di-n-propylamine | 2B | YG7108 | NT | 25 | 2.5 | 1.0 | |||||||
| [50] | N-Nitroso-di-n-butylamine | 2B | YG7108 | NT | 52 | 0.52 | ± | |||||||
| [51] | N-Nitrosomorpholine | 2B | YG7108 | NT | 0.56 | 0.63 | 0.33 | |||||||
| Vinyl compounds | ||||||||||||||
| [52] | Acrylonitrile | 2B | TA100 | NT | 0.0093 | 0.0082 | 0.0088 | |||||||
| [53] | Acrylamide | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [54] | Styrene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [55] | Safrole | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| Miscellaneous nitrogen compounds | ||||||||||||||
| [56] | Hydrazine 2HCl | 2B | TA100 | 0 | 0.16 | 0.18 | 0.10 | |||||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 2A | TA100 | ± | ± | ± | ± | |||||||
| [58] | Dacarbazine | 2B | TA100 | 1.0 | 0.55 | 0.93 | 1.1 | |||||||
| [59] | Sodium nitrite | nc | TA100 | 0.065 | 0.051 | 0.052 | 0.056 | |||||||
| [60] | Sodium nitrate | nc | TA100 | 0 | 0 | 0 | 0 | |||||||
Classification according to the Lists of IARC Evaluations (IARC Monographs home page, updated July 22, 2004): group 1, human carcinogens; group 2A, probable human carcinogens; 2B, possible human carcinogens; group 3, unclassifiable; and nc, not classified.
NT, not tested.
±, equivocal response.
Data are taken from Hakura et al. (28).
Summary data of the Salmonella/human S9 mutagenicity test
| Chemical no. . | Chemical name . | IARC groupa . | Bacterial strain . | Mutagenic activity (induced revertants/µg/plate) . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | . | -S9 mix . | Induced rat S9 . | Human S9 (HLS-014) . | Human S9 (pooled) . | |||||||
| Polycyclic hydrocarbons | ||||||||||||||
| [1] | Phenanthrene | 3 | TA100 | 0 | 0 | 0 | 0 | |||||||
| [2] | Pyrene | 3 | TA100 | NTb | 0 | 0 | 0 | |||||||
| [3] | Chrysene | 3 | TA100 | NT | 110 | ±c | 0 | |||||||
| [4] | Benz[a]anthracene | 2A | TA100 | NT | 23 | 0 | 0 | |||||||
| [5] | Dibenz[a,c]anthracene | 3 | TA100 | NT | 530 | 0 | 0 | |||||||
| [6] | Dibenz[a,h]anthracene | 2A | TA100 | NT | 2.6 | 0 | 0 | |||||||
| [7]d | Benzo[a]pyrene | 2A | TA100 | NT | 322 | 226 | ± | |||||||
| [8] | 7,12-Dimethylbenz[a]anthracene | nc | TA100 | 11 | 190 | 480 | 51 | |||||||
| [9] | 3-Methylcholanthrene | nc | TA100 | NT | 260 | 200 | ± | |||||||
| Heterocyclic hydrocarbons | ||||||||||||||
| [10] | Quinoline | nc | TA100 | 0 | 3.0 | 1.6 | 0.83 | |||||||
| [11] | 1,7-Phenanthroline | nc | TA100 | NT | 12 | 0 | 0 | |||||||
| [12] | 4-Methylquinoline | nc | TA100 | 0 | 14 | 15 | 3.0 | |||||||
| [13] | 7,9-Dimethylbenz[c]acridine | nc | TA100 | NT | 150 | 630 | ± | |||||||
| [14] | Aflatoxin B1 | 1 | TA100 | 97 | 67000 | 4000 | 810 | |||||||
| [15] | Quercetin | 3 | TA100 | NT | 36 | 40 | 37 | |||||||
| Aromatic amines | ||||||||||||||
| [16] | 2,4-Diaminotoluene | 2B | TA100 | NT | 0.044 | ± | 0 | |||||||
| [17] | o-Tolidine | 2B | TA98 | 0 | 0.3 | 0 | 0 | |||||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 2B | TA98 | NT | 210 | 160 | 27 | |||||||
| [19] | 4,4′-Methylenedianiline | 2B | TA100 | 0 | 5.1 | 4.6 | 0.11 | |||||||
| [20] | Auramine (Basic Yellow 2) | 2B | TA100 | 0 | 0 | 0 | 0 | |||||||
| [21] | 1-Naphthylamine HCl | 3 | TA100 | 0 | ± | 10 | 1.8 | |||||||
| [22] | 2-Aminofluorene | nc | TA100 | ± | 200 | 5000 | 200 | |||||||
| [23] | 2-Acetylaminofluorene | nc | TA100 | 0 | 59 | 390 | 29 | |||||||
| [24] | 2-Aminoanthracene | nc | TA100 | 0 | 1500 | 24000 | 22000 | |||||||
| [25] | 6-Aminochrysene | nc | TA100 | 21 | 4000 | 5000 | 500 | |||||||
| [26] | 1-Aminopyrene | nc | TA100 | NT | 390 | 190 | ± | |||||||
| Heterocyclic amines | ||||||||||||||
| [27] | PhIP HCl | 2B | TA100 | NT | 260 | 35 | 0.32 | |||||||
| [28] | Trp-P-2 acetate | 2B | TA100 | NT | 1400 | ± | ± | |||||||
| [29] | MeAαC acetate | 2B | TA100 | NT | 260 | 56 | 5.6 | |||||||
| [30] | IQ | 2A | TA100 | NT | 18000 | 6000 | 340 | |||||||
| [31] | MeIQx | 2B | TA100 | NT | 4000 | 2200 | 84 | |||||||
| Aromatic nitro compounds | ||||||||||||||
| [32] | Chloramphenicol | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [33] | Metronidazole | 2B | TA100 | 5.7 | 6.0 | 4.8 | 5.0 | |||||||
| [34] | 2,4-Dinitrotoluene | 2B | TA100 | 0.58 | 0.46 | 0.33 | 0.28 | |||||||
| [35] | 2-Nitrofluorene | 2B | TA100 | 560 | 430 | 460 | 800 | |||||||
| [36] | 1-Nitronaphthalene | 3 | TA100 | 34 | 19 | 58 | 80 | |||||||
| [37] | Azathioprine | 1 | TA100 | 14 | 4.3 | 3.1 | 2.2 | |||||||
| [38] | AF-2 | 2B | TA100 | 79000 | 15000 | 31000 | 41000 | |||||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | nc | TA100 | 8400 | 380 | 3200 | 3700 | |||||||
| [40] | 1-Nitropyrene | 2B | TA100 | 1000 | 280 | 1400 | 950 | |||||||
| Azo dyes | ||||||||||||||
| [41] | Azobenzene | 3 | TA100 | NT | 5.1 | 2.9 | 0 | |||||||
| [42] | 4-Aminoazobenzene | 2B | TA100 | NT | 4.6 | 1.4 | 0 | |||||||
| [43] | o-Aminoazotoluene | 2B | TA100 | 0 | 69 | 25 | ± | |||||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [45] | Congo red (Direct Red 28) | nc | TA100 | NT | 0 | 0 | 0 | |||||||
| [46a] | Trypan blue | 2B | TA98 | NT | 0 | 0 | 0 | |||||||
| [46b] | Trypan blue (crude) | 2B | TA98 | 0 | 0.14 | 1.0 | 0.49 | |||||||
| Nitrosamines | ||||||||||||||
| [47]d | N-Nitrosodimethylamine | 2A | YG7108 | NT | 8.0 | 66 | 63 | |||||||
| [48] | N-Nitrosodiethylamine | 2A | YG7108 | ± | 4.3 | 11 | 6.4 | |||||||
| [49] | N-Nitroso-di-n-propylamine | 2B | YG7108 | NT | 25 | 2.5 | 1.0 | |||||||
| [50] | N-Nitroso-di-n-butylamine | 2B | YG7108 | NT | 52 | 0.52 | ± | |||||||
| [51] | N-Nitrosomorpholine | 2B | YG7108 | NT | 0.56 | 0.63 | 0.33 | |||||||
| Vinyl compounds | ||||||||||||||
| [52] | Acrylonitrile | 2B | TA100 | NT | 0.0093 | 0.0082 | 0.0088 | |||||||
| [53] | Acrylamide | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [54] | Styrene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [55] | Safrole | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| Miscellaneous nitrogen compounds | ||||||||||||||
| [56] | Hydrazine 2HCl | 2B | TA100 | 0 | 0.16 | 0.18 | 0.10 | |||||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 2A | TA100 | ± | ± | ± | ± | |||||||
| [58] | Dacarbazine | 2B | TA100 | 1.0 | 0.55 | 0.93 | 1.1 | |||||||
| [59] | Sodium nitrite | nc | TA100 | 0.065 | 0.051 | 0.052 | 0.056 | |||||||
| [60] | Sodium nitrate | nc | TA100 | 0 | 0 | 0 | 0 | |||||||
| Chemical no. . | Chemical name . | IARC groupa . | Bacterial strain . | Mutagenic activity (induced revertants/µg/plate) . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | . | -S9 mix . | Induced rat S9 . | Human S9 (HLS-014) . | Human S9 (pooled) . | |||||||
| Polycyclic hydrocarbons | ||||||||||||||
| [1] | Phenanthrene | 3 | TA100 | 0 | 0 | 0 | 0 | |||||||
| [2] | Pyrene | 3 | TA100 | NTb | 0 | 0 | 0 | |||||||
| [3] | Chrysene | 3 | TA100 | NT | 110 | ±c | 0 | |||||||
| [4] | Benz[a]anthracene | 2A | TA100 | NT | 23 | 0 | 0 | |||||||
| [5] | Dibenz[a,c]anthracene | 3 | TA100 | NT | 530 | 0 | 0 | |||||||
| [6] | Dibenz[a,h]anthracene | 2A | TA100 | NT | 2.6 | 0 | 0 | |||||||
| [7]d | Benzo[a]pyrene | 2A | TA100 | NT | 322 | 226 | ± | |||||||
| [8] | 7,12-Dimethylbenz[a]anthracene | nc | TA100 | 11 | 190 | 480 | 51 | |||||||
| [9] | 3-Methylcholanthrene | nc | TA100 | NT | 260 | 200 | ± | |||||||
| Heterocyclic hydrocarbons | ||||||||||||||
| [10] | Quinoline | nc | TA100 | 0 | 3.0 | 1.6 | 0.83 | |||||||
| [11] | 1,7-Phenanthroline | nc | TA100 | NT | 12 | 0 | 0 | |||||||
| [12] | 4-Methylquinoline | nc | TA100 | 0 | 14 | 15 | 3.0 | |||||||
| [13] | 7,9-Dimethylbenz[c]acridine | nc | TA100 | NT | 150 | 630 | ± | |||||||
| [14] | Aflatoxin B1 | 1 | TA100 | 97 | 67000 | 4000 | 810 | |||||||
| [15] | Quercetin | 3 | TA100 | NT | 36 | 40 | 37 | |||||||
| Aromatic amines | ||||||||||||||
| [16] | 2,4-Diaminotoluene | 2B | TA100 | NT | 0.044 | ± | 0 | |||||||
| [17] | o-Tolidine | 2B | TA98 | 0 | 0.3 | 0 | 0 | |||||||
| [18] | 3,3′-Dichlorobenzidine 2HCl | 2B | TA98 | NT | 210 | 160 | 27 | |||||||
| [19] | 4,4′-Methylenedianiline | 2B | TA100 | 0 | 5.1 | 4.6 | 0.11 | |||||||
| [20] | Auramine (Basic Yellow 2) | 2B | TA100 | 0 | 0 | 0 | 0 | |||||||
| [21] | 1-Naphthylamine HCl | 3 | TA100 | 0 | ± | 10 | 1.8 | |||||||
| [22] | 2-Aminofluorene | nc | TA100 | ± | 200 | 5000 | 200 | |||||||
| [23] | 2-Acetylaminofluorene | nc | TA100 | 0 | 59 | 390 | 29 | |||||||
| [24] | 2-Aminoanthracene | nc | TA100 | 0 | 1500 | 24000 | 22000 | |||||||
| [25] | 6-Aminochrysene | nc | TA100 | 21 | 4000 | 5000 | 500 | |||||||
| [26] | 1-Aminopyrene | nc | TA100 | NT | 390 | 190 | ± | |||||||
| Heterocyclic amines | ||||||||||||||
| [27] | PhIP HCl | 2B | TA100 | NT | 260 | 35 | 0.32 | |||||||
| [28] | Trp-P-2 acetate | 2B | TA100 | NT | 1400 | ± | ± | |||||||
| [29] | MeAαC acetate | 2B | TA100 | NT | 260 | 56 | 5.6 | |||||||
| [30] | IQ | 2A | TA100 | NT | 18000 | 6000 | 340 | |||||||
| [31] | MeIQx | 2B | TA100 | NT | 4000 | 2200 | 84 | |||||||
| Aromatic nitro compounds | ||||||||||||||
| [32] | Chloramphenicol | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [33] | Metronidazole | 2B | TA100 | 5.7 | 6.0 | 4.8 | 5.0 | |||||||
| [34] | 2,4-Dinitrotoluene | 2B | TA100 | 0.58 | 0.46 | 0.33 | 0.28 | |||||||
| [35] | 2-Nitrofluorene | 2B | TA100 | 560 | 430 | 460 | 800 | |||||||
| [36] | 1-Nitronaphthalene | 3 | TA100 | 34 | 19 | 58 | 80 | |||||||
| [37] | Azathioprine | 1 | TA100 | 14 | 4.3 | 3.1 | 2.2 | |||||||
| [38] | AF-2 | 2B | TA100 | 79000 | 15000 | 31000 | 41000 | |||||||
| [39] | 4-Nitroquinoline N-oxide (4NQO) | nc | TA100 | 8400 | 380 | 3200 | 3700 | |||||||
| [40] | 1-Nitropyrene | 2B | TA100 | 1000 | 280 | 1400 | 950 | |||||||
| Azo dyes | ||||||||||||||
| [41] | Azobenzene | 3 | TA100 | NT | 5.1 | 2.9 | 0 | |||||||
| [42] | 4-Aminoazobenzene | 2B | TA100 | NT | 4.6 | 1.4 | 0 | |||||||
| [43] | o-Aminoazotoluene | 2B | TA100 | 0 | 69 | 25 | ± | |||||||
| [44] | N,N-Dimethyl-4-aminoazobenzene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [45] | Congo red (Direct Red 28) | nc | TA100 | NT | 0 | 0 | 0 | |||||||
| [46a] | Trypan blue | 2B | TA98 | NT | 0 | 0 | 0 | |||||||
| [46b] | Trypan blue (crude) | 2B | TA98 | 0 | 0.14 | 1.0 | 0.49 | |||||||
| Nitrosamines | ||||||||||||||
| [47]d | N-Nitrosodimethylamine | 2A | YG7108 | NT | 8.0 | 66 | 63 | |||||||
| [48] | N-Nitrosodiethylamine | 2A | YG7108 | ± | 4.3 | 11 | 6.4 | |||||||
| [49] | N-Nitroso-di-n-propylamine | 2B | YG7108 | NT | 25 | 2.5 | 1.0 | |||||||
| [50] | N-Nitroso-di-n-butylamine | 2B | YG7108 | NT | 52 | 0.52 | ± | |||||||
| [51] | N-Nitrosomorpholine | 2B | YG7108 | NT | 0.56 | 0.63 | 0.33 | |||||||
| Vinyl compounds | ||||||||||||||
| [52] | Acrylonitrile | 2B | TA100 | NT | 0.0093 | 0.0082 | 0.0088 | |||||||
| [53] | Acrylamide | 2A | TA100 | 0 | 0 | 0 | 0 | |||||||
| [54] | Styrene | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| [55] | Safrole | 2B | TA100 | NT | 0 | 0 | 0 | |||||||
| Miscellaneous nitrogen compounds | ||||||||||||||
| [56] | Hydrazine 2HCl | 2B | TA100 | 0 | 0.16 | 0.18 | 0.10 | |||||||
| [57] | 1,2-Dimethylhydrazine 2HCl | 2A | TA100 | ± | ± | ± | ± | |||||||
| [58] | Dacarbazine | 2B | TA100 | 1.0 | 0.55 | 0.93 | 1.1 | |||||||
| [59] | Sodium nitrite | nc | TA100 | 0.065 | 0.051 | 0.052 | 0.056 | |||||||
| [60] | Sodium nitrate | nc | TA100 | 0 | 0 | 0 | 0 | |||||||
Classification according to the Lists of IARC Evaluations (IARC Monographs home page, updated July 22, 2004): group 1, human carcinogens; group 2A, probable human carcinogens; 2B, possible human carcinogens; group 3, unclassifiable; and nc, not classified.
NT, not tested.
±, equivocal response.
Data are taken from Hakura et al. (28).
Results and discussion
The mutagenicity of 58 chemicals was determined using the Ames test in the presence of a human liver S9 fraction with a high metabolic activity (HLS-014), a pooled human liver S9 fraction (pooled S9) or a liver S9 fraction from a male SD rat treated with PB/BF (rat S9) at an equivalent amount of S9 protein (1 mg/plate). The Ames test was conducted at the 25 participating laboratories listed in Table I, all of which were experienced at performing the Ames test.
To validate the assay systems conducted in each laboratory, the mutagenicity of benzo[a]pyrene (BP) at a dose of 27 µg/plate (positive control) and of DMSO (negative control) was assayed in TA100 with the three S9 fractions. The assay data were as follows: the mean number of revertants per plate and the standard deviation of the data obtained from the 25 laboratories were 126 ± 30 for the negative control in the presence of rat S9, 124 ± 29 for the negative control in the presence of HLS-014, 130 ± 29 for the negative control in the presence of pooled S9, 827 ± 294 for the positive control in the presence of rat S9, 796 ± 284 for the positive control in the presence of HLS-014 and 245 ± 68 for the positive control in the presence of pooled S9. The variation in the assay data was regarded to be within an acceptable range and the mean values were consistent with previously reported data (28), supporting the validity of the mutagenicity data produced by the participating laboratories.
The total P450 contents and metabolic enzyme activities of cytochrome P450s towards specific substrates for each P450 subfamily were determined in the three S9 fractions (Table III). The total P450 amounts of rat S9 and HLS-014 were 16- and 3-fold that of pooled S9, respectively. The enzyme activities of HLS-014 were 5-fold higher (CYP1A1/2 and 3A) than or equal (CYP2E1) to those of pooled S9, while rat S9 exhibited 52-fold (CYP1A1/2) and 3-fold (P450 2E1 and P450 3A) higher enzyme activities than those of pooled S9. A possible explanation for why HLS-014 had such a high level of drug-metabolizing enzyme activity is that the donor liver had been subjected to enzyme induction by anti-asthma agents for a period of 10 years. Another possible reason is a high enzyme activity arising from genetic polymorphisms in drug-metabolizing enzymes. Figure 1 shows the dose–response curves for the mutagenicity of each chemical tested in this study and Table IV lists the mutagenic activity (equal to the number of induced revertants/µg/plate), as calculated from the dose–response curves shown in Figure 1 or taken from our previous study (28). Further we should like to add that the S9-mediated mutation assay used in this study would be virtually dependent on the ability of the enzymes (including no phase II enzyme) requiring NADPH/NADH as a cofactor(s).
Table IV and Figure 1 demonstrate the large diversity in the mutagenicity of chemicals in the presence of rat or human liver S9 fractions. The diversity was dependent on the chemical's structure, and a large diversity was observed particularly for compounds that probably require CYP1A1/2 for metabolic activation like polycyclic hydrocarbons and aromatic or heterocyclic amines (5,6). For example, most of the polycyclic hydrocarbons that were moderately mutagenic with the rat S9 fraction showed no mutagenicity or an equivocal response in the presence of the human S9 fractions.
For most of the test compounds that were judged to be mutagenic in the presence of either the rat or human S9 fractions (75%; 36 out of 48 compounds), the order of magnitude of the mutagenic activity of the test compounds was rat S9 ≥ HLS-014 ≥ pooled S9, as shown in Table IV. For the remaining compounds that were judged to be mutagenic (25%; 12 out of 48 chemicals), however, the mutagenic activity was more potent in the presence of both or either one of the human S9 fractions than in the rat S9 fraction: HLS-014 ≥ rat S9 ≥ pooled S9 for four mutagens [4 out of 48 chemicals (8%)]; 2 three-ring aromatic amines [2-aminofluorene and 2-acetylaminofluorene] and 2 methyl group-derivatives of polycyclic or heterocyclic hydrocarbons [7,12-dimethylbenz[a]anthracene and 7,9-dimethylbenz[c]acridine] and HLS-014 ≥ pooled S9 ≥ rat S9 for 8 mutagens [8 out of 48 chemicals (17%)]; 2 two- or three-ring aromatic amines [1-naphthylamine and 2-aminoanthracene], 4 aromatic nitro compounds [1-nitronaphthalene, AF-2, 4NQO, and 1-nitropyrene] and dimethyl- and diethyl-nitrosamines. Although all of the aromatic nitro compounds were mutagenic without the S9 mix, except for chloramphenicol, we determined their mutagenicity in the presence of the three S9 fractions to examine the effects of these S9 fractions on the mutagenicity. The order of magnitude of the mutagenic activity of the aromatic nitro compounds was without S9 ≥ pooled S9 = HLS-014 ≥ rat S9 (Table IV). A reduction in the amount of promutagens through their detoxification, such as for hydroxylated metabolites (35,36), is one possible reason for the decrease in the mutagenicity seen in the presence of the human or rat S9 fractions.
Previously, we suggested that the use of both a selected human liver S9 fraction with a relatively high level of P450-catalyzed drug metabolizing enzyme activity, when compared with the activities of several available donated livers (e.g. HLS-014), and a pooled human liver S9 fraction could be used as a possible indicator to examine inter-individual variability in mutagenic response to chemicals (28). The former and latter S9 fractions may act as indicators of mutagenic risk in humans with a high susceptibility and in humans with intermediate susceptibility to mutagens, respectively. Table IV shows that almost all the compounds tested were equally or more potently mutagenic in the presence of HLS-014 than in the presence of pooled S9. A large difference in the mutagenic activity of the chemicals in the presence of HLS-014 or pooled S9 was observed, depending upon the specific compound. In addition, negative or equivocal responses were often observed in the presence of pooled S9. If a compound exhibited a higher mutagenicity in the presence of HLS-014 than in the presence of pooled S9, this compound was considered to be characterized by large inter-individual differences in mutagenic response. These compounds included polycyclic hydrocarbons, heterocyclic hydrocarbons, aromatic amines and heterocyclic amines, which probably require CYP1A1/2 for metabolic activation (5,6). On the other hand, if the compounds showed equal mutagenicity in the presence of either HLS-014 or pooled S9, these compounds were regarded to be characterized by little or no inter-individual differences in mutagenic response.
No significant correlations were observed between the mutagenic activities (the number of induced revertants/µg/plate) of the chemicals induced by each S9 fraction (Table IV) and each enzyme activity of the P450 subfamily (Table III). This finding supports the idea that the mutagenic activity of the chemicals is determined not only by the single P450 enzyme activity involved in its metabolic activation but by multiple enzymatic activation pathways or the balance between metabolic activation and detoxification (12,25,28). Therefore, the use of the S9 fraction may be valuable as a convenient system for the comprehensive evaluation of mutagenicity in humans, compared with genetically engineered test strains expressing human cytochrome P450s, which may be useful for determining the contribution of each P450 subfamily to the metabolic activation or detoxification of promutagens (37–40). The microsome fraction may also be useful as a simpler, mechanistic evaluation system (13,41–46).
As mentioned above, a large diversity in the mutagenicity of the chemicals in the presence of the rat and human liver S9 fractions was observed. What are the reasons for this diversity? One reason is the difference in the quantity of the S9 fraction, or the different amounts of drug-metabolizing enzymes (including P450s), between the S9 fractions (Table III). Another probable reason is the diversity in the quality of the S9 fractions; the mutagenic activity per total P450 content (induced revertants/µg/plate/total P450 content) varies between the rat and human S9 fractions by as much as 100-fold, depending on the chemical (data to be published elsewhere). This finding also suggests that the use of human S9 fractions may be advantageous.
The mutagenicity of 3-methylcholanthrene and 3,3′-dichlorobenzidine HCl was determined in TA100 and TA98 strains in the presence of rat S9, HLS-014 or pooled S9 (Figure 2). These compounds showed a different mutagenic response to TA100 and TA98 in the presence of each S9 fraction. This finding indicates that each S9 fraction produces a different ratio of characteristic DNA damage leading to mutation in TA100 (base pair-substitution type) or TA98 (frameshift type). For example, 3,3′-dichlorobenzidine HCl is suggested to preferentially produce DNA injury causing frameshift mutations rather than damage causing base pair-substitution mutations in the presence of the human S9 fractions, compared with what happened in the presence of the rat S9 fraction. This observation may reflect the diversity in the amount, substrate specificity, and metabolic rate of each enzyme involved in mutagenic activation and detoxification among each S9 fraction. This additional evidence also shows the diversity in the quality of the S9 fractions, supporting the advantage of using human S9 fractions in mutagenicity testing systems.
Comparison of mutagenic activities in the presence of the human and rat S9 fractions between TA100 and TA98. The mutagenicity of 3-methylcholanthrene [(a) and (b)] and 3,3′-dichlorobenzidine 2HCl [(c) and (d)] towards TA100 [(a) and (c)] and TA98 [(b) and (d)] was determined in the presence of the HLS-014, pooled S9 or rat S9 fractions. The symbols closed square (▪), open circle (○), and closed circle (•) used in the dose–response curves indicate rat S9, HLS-014, and pooled S9, respectively.
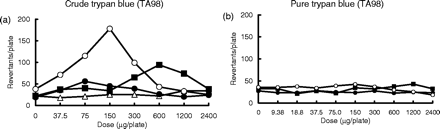
A pure test article of trypan blue, an azo dye compound, was non-mutagenic in the presence of the human S9 and rat S9 fractions, while a crude one (manufacturers' guaranteed reagent) was mutagenic in the presence of these S9 fractions; the most potently mutagenic activity in the presence of HLS-014, the second most potently mutagenic activity was observed in the presence of pooled S9, and the most weekly mutagenic activity was observed in the presence of rat S9 (Figure 3 and Table IV). The crude test article of trypan blue tested in this study contained some aromatic amines as impurities, and the mutagenic response of the test strain to the crude test article in the presence of the three S9 fractions was similar to that found for the aromatic amines, demonstrated in Figure 1 and Table IV. Based on these results, the mutagenicity of the crude test article of trypan blue may be due to the aromatic amines present, although a previous report has stated that the mutagenicity of trypan blue purchased from Eastman is, partially, most likely due to impurities such as azo compounds that require reduction (31,47). As previously mentioned, some mutagens including aromatic amines, were more mutagenic in the presence of the human S9 than in the presence of the rat S9 fraction. This finding is of great importance for evaluations of genotoxicity of chemicals in humans, again suggesting that the use of human S9 fractions may be advantageous.
Mutagenicity of the crude and pure test articles of trypan blue. The mutagenicity of crude (a) and pure (b) trypan blue test articles towards TA98 was determined in the presence and absence of the rat S9, HLS-014 or pooled S9 fractions. The crude test article of trypan blue tested was purchased from Nacalai as a manufacturer's guaranteed reagent, and its purity was roughly estimated to be 50%, although it was not specifically determined. On the other hand, the purity of the pure trypan blue test article was 96%. The symbols open triangle (▵), closed square (▪), open circle (○), and closed circle (•) used in the dose–response curves indicate treatments without S9 mix, with rat S9, with HLS-014 and with pooled S9, respectively.
Test compounds that were non-mutagenic in the presence of the rat S9 fraction (for example, the vinyl compounds) were also non-mutagenic in the presence of human S9 fractions. A few reports have demonstrated that some halogenated vinyl compounds, thought to be activated for mutagenicity through the epoxidation of their vinyl group (4), were less mutagenic in the presence of human S9 than in the presence of induced rat S9 (11). Hence, vinyl compounds are unlikely to be activated by human S9 as efficiently as they are by induced rat S9 under the in vitro conditions employed.
The use of rodent S9 fractions with or without pretreatment with drug-metabolizing inducer(s) in mutagenicity testing systems is advantageous for screening programs to identify hazardous chemicals for the following reasons: (i) they effectively activate promutagens to mutagens, (ii) they are convenient, (iii) they are cost-effective and (iv) they produce relatively consistent responses to test substances, which may be important in regulatory decision-making (2). In spite of these advantages, the use of human S9 fractions may also be rational because they reflect the human metabolic system more faithfully. In fact, numerous studies have been conducted on variations in metabolism between animals and humans (48,49), and human S9 or microsome fractions have been frequently used to predict the metabolism of drugs in humans in recent years. Furthermore, variations in metabolism produce quantitative and qualitative differences in mutagenicity between rat and human S9 fractions, as demonstrated in this study. Therefore, we believe that assays utilizing human S9 fractions are advantageous as in vitro human model systems to improve our understanding of the mutagenic effects of chemicals on humans. The classification of carcinogens by the IARC is presently based on weight-of-evidence supporting carcinogenicity in humans. Mutagenicity tests using human S9 fractions may become a valuable tool in this process. Human S9 is a simulated model and an aspect of human metabolism (50); hence, mutagenicity tests using human cells coupled with human S9 may be more valuable than the Ames test coupled with human S9, in terms of a more proximate in vitro human model. Human S9 fractions prepared from individual or pooled donor tissues are both commercially available from HAB and some companies (e.g. XenoTech and In Vitro Technologies). However, three important disadvantages of using human S9 must be resolved: (i) human S9 is relatively expensive, (ii) most of the human S9 fractions presently available are derived from Caucasian donors and (iii) there is no consensus on the human S9 fractions to be used as standards for evaluating mutagenicity because of the heterogeneous nature of humans (25,27–29).
In conclusion, we strongly suggest that induced rat S9 fractions be routinely used as a first choice for hazard identification and that human S9 fractions may be used in comprehensive assessments of mutagenicity as second-tier assays. Furthermore, a conventional method that utilizes pooled S9 and a selected human S9 fraction with a potent drug-metabolizing activity may be useful for preliminary evaluations of inter-individual differences in the mutagenicity of chemicals between humans, although multiple S9 fractions would be ideal.
Presented in part at the 8th International Conference on Environmental Mutagens, Shizuoka, Japan, 2001.
References
Maron,D.M. and Ames,B.N. (
Mortelmans,K. and Zeiger,E. (
Ames,B.N., Durston,W.E., Yamasaki,E. and Lee,F.D. (
Cooper,C.S. and Grover,P.L. (
Guengerich,F.P. and Shimada,T. (
Callander,R.D., Mackay,J.M., Clay,P., Elcombe,C.R. and Elliott,B.M. (
Franco,S.G., Domínguez,G. and Pico,J.C. (
Tang,T. and Friedman,M.A. (
Buening,M.K., Fortner,J.G., Kappas,A. and Conney,A.H. (
Bartsch,H., Malaveille,C., Barbin,A. and Planche,G. (
Dybing,E., Bahr,C.v., Aune,T., Glaumann,H., Levitt,D.S. and Thorgeirsson,S.S. (
Sabadie,N., Malaveille,C., Camus,A-M. and Bartsch,H. (
Phillipson,C.E. and Ioannides,C. (
Phillipson,C.E. and Ioannides,C. (
Phillipson,C.E. and Ioannides,C. (
Beaune,P., Lemestré-Cornet,R., Kremers,P., Albert,A. and Gielen,J. (
Le,J., Jung,R. and Kramer,M. (
Mori,Y., Yamazaki,H., Toyoshi,K., Maruyama,H. and Konishi,Y. (
Neis,J.M., Yap,S.H., van Gemert,P.J.L., Roelofs,H.M.J., Bos,R.P. and Henderson,P.Th. (
Raineri,R., Andrews,A.W. and Poiley,J.A. (
Yamazaki,H., Mori,Y., Toyoshi,K., Nagai,H., Koda,A. and Konishi,Y. (
Jongeneelen,F.J., Akker,W.v.d., Bos,R.P., Anzion,R.B.M., Theuws,J.L.G., Roelofs,H.M.J. and Henderson,P.Th. (
Smith,A.J. and Chipman,J.K. (
Johnson,T.E., Umbenhauer,D.R. and Galloway,S.M. (
Hakura,A., Suzuki,S. and Satoh,T. (
Hakura,A., Suzuki,S., Sawada,S., Motooka,S. and Satoh,T. (
Hakura,A., Suzuki,S., Sawada,S., Sugihara,T., Hori,Y., Uchida,K., Kerns,W.D., Sagami, F., Motooka,S. and Satoh,T. (
Hakura,A., Suzuki,S. and Satoh,T. (
Lijinsky,W. and Andrews,A.W. (
Prival,M.J., Bell,S.J., Mitchell,V.D., Peiperl,M.D. and Vaughan,V.L. (
Yamada,M., Matsui,K., Sofuni,T. and Nohmi,T. (
Omura,T. and Sato,R. (
Ikeda,T., Nishimura,K., Taniguchi,T. et al. (
Synder,S., Hsu,I.C. and Trump,B.F. (
Kataoka,K., Kinouchi,T. and Ohnishi,Y. (
Cooper,M.T. and Porter,T.D. (
Kushida,H., Fujita,K., Suzuki,A., Yamada,M., Endo,T., Nohmi,T. and Kamataki,T. (
Fujita,K. and Kamataki,T. (
Josephy,P.D., Batty,S.M. and Boverhof,D.R. (
Czygan,P., Greim,H., Garro,A.J., Hutterer,F., Rudick,J., Schaffner,F. and Popper,H. (
Boobis,A.R., Brodie,M.J., McManus,M.E., Staiano,N., Thorgeirsson,S.S., and Davies,D.S. (
Yamazoe,Y., Abu-Zeid,M., Yamauchi,K. and Kato,R. (
Kitada,M., Taneda,M., Ohta,K., Nagashima,K., Itahashi,K. and Kamataki,T. (
Marczylo,T. and Ioannides,C. (
Yamada,K., Suzuki,T., Hakura,A., Mizutani,T. and Saeki, K. (
Prival,M.J. and Mitchell,V.D. (
Distlerath,L.M. and Guengerich,F.P. (
Shimada,T. and Okuda,Y. (
Author notes
Drug Safety Research Laboratories, Eisai Co., Ltd, 1 Takehaya-machi, Kawashima-cho, Hashima-gun, Gifu 501-6195, Japan, 1Drug Safety Research Laboratory, Daiichi Pharmaceutical Co., Ltd, 16-13 Kita-Kasai 1-Chome, Edogawa-ku, Tokyo 134-8630, Japan, 2Biosafety Research Center, Foods, Drugs and Pesticides, 582-2 Shioshinden, Fukude-cho, Iwata-gun, Shizuoka 437-1213, Japan, 3Food and Drug Safety Center, 729-5 Ochiai, Hadano, Kanagawa 257-8523, Japan, 4Environmental Health Science Laboratory, Sumitomo Chemical Co., Ltd, 3-1-98 Kasugadenaka, Konohana-ku, Osaka 554-8558, Japan and 5HAB Biomedical Research Institute, Cornea Center Building, Ichikawa General Hospital, 5-11-13 Sugano, Ichikawa, Chiba 272-8513, Japan



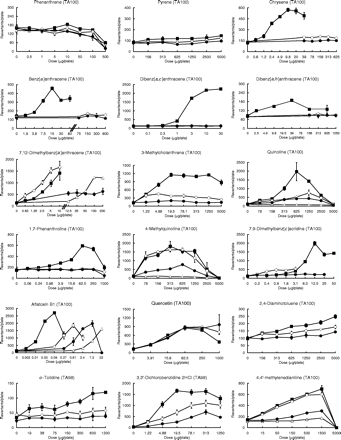
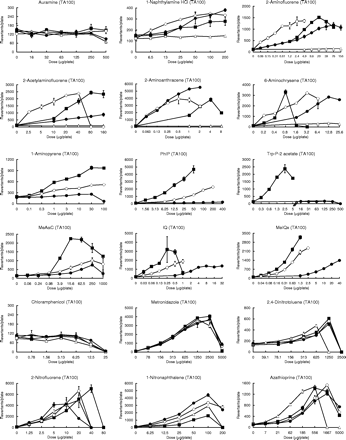
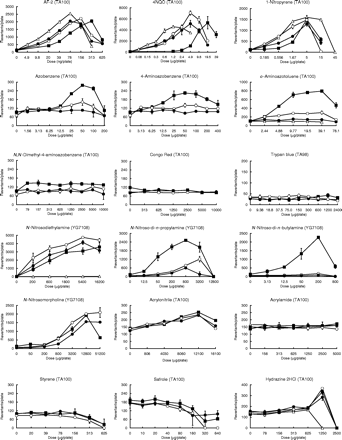
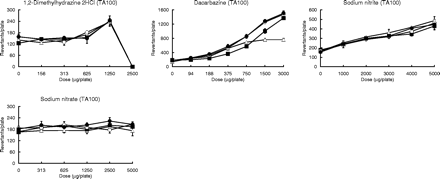
![Comparison of mutagenic activities in the presence of the human and rat S9 fractions between TA100 and TA98. The mutagenicity of 3-methylcholanthrene [(a) and (b)] and 3,3′-dichlorobenzidine 2HCl [(c) and (d)] towards TA100 [(a) and (c)] and TA98 [(b) and (d)] was determined in the presence of the HLS-014, pooled S9 or rat S9 fractions. The symbols closed square (▪), open circle (○), and closed circle (•) used in the dose–response curves indicate rat S9, HLS-014, and pooled S9, respectively.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/mutage/20/3/10.1093/mutage/gei029/2/m_gei029f2.gif?Expires=1716341019&Signature=3kqzfG9OqYZtrMC7eaalg66PRB2fJiygJ8TCASQtZY~DLHJ52DgOFPblMFtRKIsrJji5TOQA7zoNh~NkGCONU3XWc5EAFf9rLrExcw8onpy0i6OVkA6RjHaFrmYXcTKzrys8cWsz6sEj~ky99Ie23NSvgOpeAITQAzYmKtxVgj0F~mUNb1j57PzWllXDnr~LPCyJUgvWavKN4pr3OJ0oooCLR4t4o371-O6RFHUUjkO~xgqGs5TwIS5~UMt0vTiaEdlO4eCd-9tzXDzrF7US9S~FsAG5GFqQbj9kJBPafYukXgKtt6TDENVwhb9UmCFmlK6j58qmM-uHrHz9mPDkOA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)