Abstract
Considering the fatal human victims and economic loss caused by influenza virus infection every year, methodologies for rapid and on-site detection of influenza viruses are urgently needed. LAMP is the most commonly used nucleic acid isothermal amplification technology suitable for on-site use. However, for multiplex LAMP, differentiation of the amplicons derived from multiple targets is still challengeable currently. Here we developed a multiplex RT-LAMP assay for simultaneous amplification of three prominent subtypes of influenza viruses (A/H5, A/H7 and 2009A/H1). The amplicons were further identified by cascade invasive reaction and nanoparticle hybridization in separate target-specific detection tubes (referred to as mRT-LAMP-IRNH). The analytic sensitivities of the assay are 10 copies of RNA for all the three HA subtypes, and the specificity reached 100%. Clinical specimen analysis showed this assay had a combined sensitivity and specificity of 98.1% and 100%, respectively. Overall, the mRT-LAMP-IRNH assay can be used as a cost-saving method that utilizes a simple instrument to detect A/H5, A/H7, and 2009A/H1 influenza viruses, especially in resource-limited settings.
Similar content being viewed by others
Introduction
Influenza viruses are the major pathogens causing human respiratory diseases with severe morbidity and mortality worldwide1,2. Most influenza pandemics are associated with type A influenza viruses which can be further subdivided into subtypes based on the two types of viral surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA)3. Among viruses isolated from aquatic birds, 16 HA and 9 NA subtypes have been identified to date3, although only a small subset (H1N1, H2N2, and H3N2) have acquired the capacity to cross species barriers and subsequently establish lineages in humans over the past century4. By antigenic drift and shift, influenza viruses evolve rapidly. The predominant seasonal influenza virus strain is 2009A/H1N1 in recent years, the cause of the 2009 H1N1 pandemic5,6. In addition, human infection of various subtypes of avian-origin influenza A viruses, especially H5 subtype (H5N1, H5N6) and H7 subtype (H7N7, H7N3, H7N9), has often been reported over the past few years7,8,9,10.
The rapid detection of these viruses is crucial for epidemiologic investigations and timely responses to an influenza pandemic threat. Molecular diagnostic methods based on nucleic acid amplification tests are more rapid and sensitive than traditional techniques including virus isolation and serological assays. Single or multiplex RT-PCR (including real-time RT-PCR) is at present the powerful method for detection of influenza viruses11,12,13,14,15. However, it requires bulky and expensive equipments, as well as highly skilled technicians, which make these methods not suitable for use in resource limited regions or for field use. Recently, researches have focused on the development of isothermal amplification methods for pathogen detection. As the reaction is conducted under isothermal conditions, it can be carried out with a simple water bath so that a thermal cycler is not required16. As the most commonly used isothermal amplification method at present, loop-mediated isothermal amplification (LAMP) method is rapid and sensitive for amplification of DNA at a constant temperature of 60–65 °C17,18. Besides targeting DNA templates, LAMP can also be used to amplify RNA template by the use of reverse transcriptase together with DNA polymerase, so-called reverse transcriptase LAMP (RT-LAMP). RT-LAMP methods have been developed to detect various RNA pathogens including influenza viruses19,20,21,22,23.
Simultaneous detection of multiple pathogens in one tube is without doubt cost- and time- saving. However, the differentiation of the ladder-like LAMP amplicons derived from multiple targets is still challengeable to date, although previous studies have described several methods for multiplex LAMP detection. These multiplex LAMP methods either used end point analysis, through gel electrophoresis24 or pyrosequencing25, or used real-time detection, through annealing curve analysis26, DARQ27 or MERT-LAMP technique28. However, these techniques all require complicated and specialized instrumentations, which diminish the point-of-care testing capability of multiplex LAMP.
Because the aggregation of gold nanoparticles (AuNPs) can cause the change of their optical property, AuNPs have been used as a sensor for DNA detection29,30,31. The main merit of this sensor is the visible detection by naked eyes, and thus especially suitable for field use. In this study, we developed a multiplex RT-LAMP assay for simultaneous amplification of three prominent subtypes of influenza A viruses (H5, H7 subtypes of avian influenza A and 2009A/H1N1 viruses), and the multiplex RT-LAMP amplicons were further identified by cascade invasive reaction32,33 and gold nanoparticle hybridization in separate target-specific detection tubes. Multiplex RT-LAMP coupled with cascade invasive reaction and nanoparticle hybridization (termed as mRT-LAMP-IRNH in brief) provides us a sensitive, specific and cost-saving diagnostic tool for identification of influenza A viruses, especially in resource-limited situations.
Results
Development and optimization of the mRT-LAMP-IRNH assay
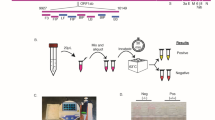
At the commencement of this study, we used a real-time turbidimeter enabling observation of primer kinetics to determine the optimal primer sequences, primer concentrations, incubation temperature, as well as incubation time of the multiplex RT-LAMP reaction. The results showed that the optimal multiplex RT-LAMP reaction was obtained with primer sequences shown in Table 1 and the primer concentrations described in Materials and Methods. At 63 °C, the best amplification efficiency was observed for all the three HA subtypes (A/H5, A/H7 and 2009A/H1), and thus 63 °C was considered to be optimal temperature for the multiplex RT-LAMP reaction (data not shown). According to the amplification curves obtained by real-time turbidimeter, 50 min was selected as the optimal incubation time. In mRT-LAMP-IRNH, the multiplex RT-LAMP amplicons were further identified by cascade invasive reaction and nanoparticle hybridization in separate target-specific detection tubes. A schematic of the principles of cascade invasive reaction and gold nanoparticle hybridization is shown in Fig. 1. In the positive reaction, the cleaved hairpin probe cannot trigger the aggregation of AuNPs, leading to a red color of the reaction. Conversely, in the negative reaction, the intact hairpin probe leads to the aggregation of AuNPs, and then the reaction mixture becomes colorless (Fig. 1).
Step 1 (Primary invasive reaction): A target DNA is firstly hybridized with an upstream probe (Up) and a downstream probe (Dp), forming a one-base overlapping structure at the 3′-end of the Up. The blue arrow indicates the site of cleavage. Step 2 (Secondary invasive reaction): The cleaved flaps from the target-specific primary invasive reaction are used to drive a secondary invasive reaction. Then the hairpin probe (Hp) is cleaved by AfuFEN. Step 3 (Nanopartical hybridization): When the target is present, the cleaved HP cannot trigger the aggregation of AuNPs, leading to a red color of the reaction. Conversely, when the target is absent, the Hp is intact, leading to the aggregation of AuNPs, and then the reaction becomes colorless.
Analytical sensitivity of the mRT-LAMP-IRNH
The analytical sensitivity of the mRT-LAMP-IRNH assay was determined by testing ten-fold serial dilutions of viral RNA transcripts (ranging from 104 to100 RNA copies) on two separate days. After 50 min of amplification, diluted RT-LAMP products were added into the invasive reaction mixture to perform cascade invasive reactions for each target, then the AuNPs were added for hybridization. The detection limits for all the three HA genes (A/H5, A/H7 and 2009A/H1) were 101 copies of synthetic RNA by presenting red color in the reaction tubes as shown in Fig. 2a–c. Identical results were obtained on both days indicating that the mRT-LAMP-IRNH assay was both robust and reproducible.
A/H5 (a), A/H7 (b) or 2009A/H1 (c) in vitro RNA transcripts (ranging from 104 to100 RNA copies) were used as RT-LAMP template, respctively. The amplicons from each RT-LAMP reaction were further identified by cascade invasive reaction and gold nanoparticle hybridization in corresponding target-specific detection tube. The multiplex RT-LAMP amplification reactions for A/H5 (d), A/H7 (e), and 2009A/H1 (f) were also monitored by real-time turbidity detection and the corresponding curves of concentrations of templates were marked in the figure. DW: distilled water used as no-template control.
To further evaluate the sensitivity of cascade invasive reaction and nanoparticle hybridization (IRNH) for LAMP product detection, the multiplex RT-LAMP amplifications were also monitored by a real-time turbidimeter. As shown in Fig. 2d–f, the detection limits for A/H7 and 2009A/H1 were both 101 copies of synthetic RNA, which were equivalent to their detection limits obtained by mRT-LAMP-IRNH. However, the detection limit for A/H5 was 102 copies of synthetic RNA, which was an order of magnitude lower than that obtained by mRT-LAMP-IRNH, indicating the sensitivity of cascade invasive reaction coupled with nanoparticle hybridization was higher than real-time turbidity detection for RT-LAMP product analysis.
Analytical specificity of the mRT-LAMP-IRNH
To assess the potential for the mRT-LAMP-IRNH assay to cross-react with other genetically or clinically related viruses which could cause similar symptoms, a specificity test was conducted using viral RNA extracts from various control viruses. As shown in Fig. 3, positive reactions with red color were only observed in the preparations of A/H5N1, A/H7N9 and 2009A/H1N1 viruses, whereas none of the control viruses showed a positive result, indicating the high specificity of the mRT-LAMP-IRNH assay.
RNA extracted from influenza viruses A/H5N1, A/H7N9, 2009A/H1N1, or each control virus was used as RT-LAMP template, respctively. The amplicons from each RT-LAMP reaction were further identified by cascade invasive reaction and gold nanoparticle hybridization in three target-specific detection tubes. (a) Detection tube for A/H5. The cascade invasive reactions were performed with H5-UP, H5-DP and HP. (b) Detection tube for A/H7. The cascade invasive reactions were performed with H7-UP, H7-DP and HP. (c) Detection tube for 2009A/H1. The cascade invasive reactions were performed with 09H1-UP, 09H1-DP and HP. 1: A/H5N1; 2: A/H7N9; 3: 2009A/H1N1; 4: A/H1N1; 5: A/H3N2; 6: influenza B; 7: A/H9N2; 8: parainfluenza viruses types 1; 9: parainfluenza viruses types 2; 10: parainfluenza viruses types 3; 11: parainfluenza viruses types 4; 12: respiratory syncytial viruses types A; 13: respiratory syncytial viruses types B; 14: distilled water used as no-template control; UP: upstream probe; DP: downstream probe; HP: hairpin probe.
Clinical sample analysis by the mRT-LAMP-IRNH
To evaluate the performance characteristics of the mRT-LAMP-IRNH assay in clinical sample detection, a total of 88 clinical specimens were subjected to mRT-LAMP-IRNH assay with the parallel analysis by real-time RT-PCR. The results showed that, out of 52 specimens that were positive for influenza as detected by real-time RT-PCR, mRT-LAMP-IRNH detected 6/6A/H5, 18/18A/H7 and 27/28 2009A/H1. The sensitivity and specificity for detecting A/H5 were 100% (6/6) and 100% (82/82), respectively. For detecting A/H7, the sensitivity and specificity were also 100% (18/18) and 100% (70/70) respectively, and for 2009A/H1, the sensitivity and specificity were 96.4% (27/28) and 100% (60/60), respectively. Based on combined A/H5, A/H7 and 2009A/H1 detection, the mRT-LAMP-IRNH assay had an overall sensitivity and specificity of 98.1% (51/52) and 100% (36/36), respectively (Table 2).
Discussion
Viral culture paired with serological HA typing is the standard method for detecting and typing influenza A viruses34. The main drawbacks of virus culture are that it can only detect live viruses and requires more time and higher biosecurity. Immunological methods for testing influenza viruses have low skill requirements, but poor sensitivity and specificity. Recently, many molecular diagnostic approaches such as single or multiplex RT-PCR (including real-time RT-PCR) have been developed for subtyping influenza viruses. However, these methods require expensive thermal cycling equipments. Currently, rapid, reliable and affordable point-of-care tests for influenza virus detection are urgently needed, especially in resource limited regions. In this study, we describe a sensitive mRT-LAMP-IRNH assay for the detection of three influenza A viruses (subtypes A/H5, A/H7 and 2009A/H1) for the first time. The read-out can be observed by naked eyes, and no specialized instrument is required, which make this assay especially useful in resource-limited situations such as primary care facilities.
As LAMP belongs to isothermal amplification methods, it can be carried out with a simple water bath and without the need of bulky and expensive equipments, which points to the potential applicability of the assay for clinical point-of-care diagnostic use. LAMP products can be detected by agarose gel electrophoresis, turbidity or fluorescence detection35,36, lateral flow dipstick23, or even visual inspection37,38. These methods are robust and reliable, but detect total amplification products in a reaction and are thus limited to detection of a single target. Multiplex detection is the development trend of pathogen detection technology due to the properties of cost savings and high efficiency. In order to achieve multiplex LAMP detection, methods based on end point analysis24,25 as well as real-time detection26,27,28 have been employed to differentiate multiple target sequences, while these strategies all required complicated and specialized instruments. To enable multiplex pathogen detection without the need of specialized instruments, Dou et al. developed a polymer/paper hybrid microfluidic biochip for simultaneous LAMP detection of three meningitis-causing pathogens. Though this assay was not really a multiplex LAMP reaction, by using of microfluidic biochip, three pathogens were simultaneously detected on a chip, and high sensitivity and specificity were achieved within 1 h39. In this study, we present an alternative molecular method for multiplex LAMP amplicon detection by combining invader techniques32,33 and gold nanoparticle probe techniques29,30,31. AuNPs have been used as a sensor for DNA detection for years. To improve the sensitivity, we coupled invader techniques with gold nanoparticle probe techniques, which were further used to detect multiplex RT-LAMP amplicons for the first time. Although the identification of the multiplex RT-LAMP amplicons is monoplex, the mRT-LAMP-IRNH assay developed in this study is a cost-saving method that requires no complicated instrument, and is more suitable for multiple-target detection of limited amount and/or precious nucleic acids. This pilot study also provides a strategy for establishing multiplex LAMP assay much more than three plex for field use. Meanwhile, unknown clinical specimens with co-infection can also be distinguished without using any equipments.
Due to the use of six sequence-specific primers per target, similar to that of the ordinary LAMP technique, which recognizes eight conserved regions for specific identification of positive targets, mRT-LAMP-IRNH offers the same high specificity. Moreover, the invasive reaction was a specific DNA detection method by the use of two target-specific probes. The mRT-LAMP-IRNH assay combines the specificities of both LAMP and the cascade invasive reaction, which should provide higher specificity in theory. In this study, we demonstrated the high specificity of the assay by showing the absence of cross-reactivity by analyzing the RNA extracts from various genetically or clinically related viruses which could cause similar symptoms.
The concordance of high analytical sensitivity between LAMP and other sensitive molecular methods has been reported previously40,41. LAMP reaction was able to tolerate the inhibitory effect of large amounts of templates, and was less affected by the presence of various salts and inhibitors42. In mRT-LAMP-IRNH method, aggregation of AuNPs is achieved by adding hairpin probes complementary to both AuNPs. The cleavage of hairpin probes is triggered by the cleaved flaps from the target-specific primary invasive reaction. Because one LAMP amplicon can yield large numbers of cleaved hairpin probes, the sensitivity of this method is increased significantly. By testing ten-fold serial dilutions of viral RNA transcripts, the analytical sensitivities of the mRT-LAMP-IRNH assay were found to be 101 copies of synthetic RNA for all three targets. Previous studies have shown that although multiplex nucleic acid detection tests have the advantage of high efficiency, due to the mutual interference of multiple primers, the analytic sensitivities of these assays tend to decrease. However, through the signal amplification effects of invader techniques and gold nanoparticle probe techniques, the sensitivity of the mRT-LAMP-IRNH assay established in this study was equivalent or even significantly increased as compared to general monoplex LAMP method23,43,44,45. In this study, the detection limit for A/H5 obtained by mRT-LAMP-IRNH was ten times higher than that obtained by real-time turbidity detection, indicating the cascade invasive reaction coupled with AuNPs hybridization increased the sensitivity of LAMP whose results were usually analyzed through turbidity.
The performance characteristics of the mRT-LAMP-IRNH assay were evaluated by testing 88 clinical specimens with the parallel analysis by real-time RT-PCR. Compared to real-time RT-PCR, the sensitivities of the mRT-LAMP-IRNH assay for detecting A/H5, A/H7 and 2009A/H1 were 100%, 100% and 96.4%, respectively. As 50 min was selected for multiplex LAMP reaction, the total detection time of mRT-LAMP-IRNH assay was about 100~120 min which was similar to that of real-time RT-PCR assay. As 120 min might be a little long for field use, we had tried to shorten the invasive reaction and hybridization reaction time, but we found that when the concentration of virus was low, the result might be somewhat ambiguous to the naked eye if the detection time was shortened. In future, we would test more conditions and settings to try to shorten the reaction time, and to combine invasive reaction with hybridization reaction in one step. These would make the assay more timely and practical for field use.
In summary, a highly sensitive and specific mRT-LAMP-IRNH assay was developed for the first time in this study which can detect three prominent subtypes of influenza viruses (A/H5, A/H7 and 2009A/H1). The advantages of cost-saving and no requirement of any complicated instrument make this assay more suitable for low-equipment setting laboratory use and for on-site testing. Consequently, this detection method constructed in this study would facilitate initial clinical treatment, infection control, as well as epidemiologic investigations of influenza infection.
Methods
Ethics statement
Written informed consent for using the clinical specimens was obtained from all patients involved in this study. This research was approved by the Ethics Committee of the Jiangsu Provincial Center for Disease Prevention and Control. The methods were carried out in accordance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects.
Viral isolates and clinical specimens
Influenza virus strains (A/Nanjing/2/2013 (H7N9), A/Jiangsu/1/2007(H5N1), and A/Jiangsu/2/2009(H1N1)) isolated from patient samples at Jiangsu Provincial Center for Disease Prevention and Control were included in the mRT-LAMP-IRNH assay development. Other genetically or clinically related virus isolates including seasonal influenza viruses (A/H1N1, A/H3N2, and B), avian influenza virus A/H9N2, parainfluenza viruses (types 1, 2, 3, and 4), and respiratory syncytial viruses (types A and B) were used as control viruses to assess the specificity of the assay. For methodological evaluation of this study, a total of 84 nasopharyngeal swabs were collected from influenza-like cases. Among these specimens, 48 had been identified positive for influenza A viruses including 18A/H7N9, 2A/H5N1, and 28 2009A/H1N1, and 36 had been identified negative for influenza A viruses. Furthermore, 4 known positive poultry cage surface swabs (2A/H5N1 and 2A/H5N6) were collected from live poultry markets. All these isolates and specimens were stored at −80 °C until use. This study was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Prevention and Control.
Extraction of viral RNA
Viral RNA was extracted from 200 μl of samples with a MagNA Pure LC Total Nucleic Acid Isolation Kit on a MagNA PureTM system (Roche Diagnostics, Manheim, Germany) according to the manufacturer’s instructions. The extracted RNA was dissolved in 50 μl elution buffer (pH = 8.0) and kept at −80 °C until use.
Preparation of transcripts
The HA genes were amplified from three influenza virus strains (A/Nanjing/2/2013(H7N9), A/Jiangsu/1/2007(H5N1), and A/Jiangsu/2/2009(H1N1)) with primers containing T7 promoter sequence in the reverse sides. PCR amplicons containing the full length region of each gene target for multiplex RT-LAMP were in vitro transcribed with T7 RNA polymerase (TaKaRa Biotechnology Co. Ltd., Dalian, China) according to the manufacturer’s instructions. The synthetic RNA transcripts were purified, quantified, and then tenfold diluted ranging from 104 to100 RNA copies/μl.
Design of primers and probes
Multiplex RT-LAMP primers for avian influenza A/H5, A/H7 and 2009A/H1 were designed using conserved regions of the HA gene for each influenza A subtype, respectively. Primers were designed using the PrimerExplorer version 4 program (Eiken Chemical Co., Tokyo, Japan). The feasibility and specificity of the primers were subsequently checked by BLAST search with sequences in GenBank. The sequence information of the final selected multiplex RT-LAMP primers is shown in Table 1. The cascade invasive reaction upstream probes and downstream probes were designed using the Universal Invader Design Software version 1.2.4 (Third Wave Technologies, Inc., Madison, USA) according to the sequences of multiplex RT-LAMP amplification products. The sequence information of the final selected upstream probes, downstream probes, hairpin probes used in secondary invasive reaction, as well as the two probes used for modification of AuNPs is shown in Table 3. All the primers and probes were synthesized by TaKaRa Biotechnology Co. Ltd. (Dalian, China).
Preparation and modification of AuNPs
AuNPs were synthesized by reducing tetrachloroauric acid with trisodium citrate according to the protocol described previously46. Transmission electron micrographs were taken to measure the size of synthetic particles (an average diameter of 13 nm). Two types of oligonucleotide probes (Table 3) were used to separately modify the two batches of the AuNPs.
Multiplex RT-LAMP reaction
Multiplex RT-LAMP reaction was carried out with a RNA Amplification Kit (RT-LAMP) (Eiken China Co., Ltd., Shanghai, China). The reaction was performed in 20 μl of a mixture containing 10 μl of Reaction Mix, 0.8 μl of Enzyme Mix, 18 primers for avian influenza A/H5, A/H7and 2009A/H1 (each of the outer primers F3 and B3 at 0.15 μM, each of the inner primers FIP and BIP at 1.2 μM, and each of the loop primers LF and LB at 0.6 μM final concentration), and 1 μl of RNA template. The reaction mixture containing 1 μl of distilled water was used as no-template controls. Multiplex RT-LAMP amplification reactions were carried out at 60, 63, and 65 °C to determine the shortest amplification time and best detection performance. During assay development, including sensitivity and specificity tests, a real-time turbidimeter (LA320C, Teramecs, Tokyo, Japan) was used for RT-LAMP amplification reaction. And a heat-block (ThermoStat Plus, Eppendorf, Hamburg, Germany) was used in the analysis of clinical samples.
Cascade invasive reaction and gold nanoparticle hybridization
For each multiplex RT-LAMP reaction, three target-specific cascade invasive reactions were performed. The 20 μl invasive reaction was carried out with 0.2 μM upstream probe, 0.2 μM downstream probe, 0.2 μM hairpin probe, 100 ng AfuFEN enzyme which was prepared in our lab as described before47, and 5 μl diluted multiplex RT-LAMP amplification products (20 μl products plus 30 μl deionized water) in a reaction buffer containing 10 mM MOPS (pH7.5), 0.05% Tween-20, 0.05% Nonidet P40, and 7.5 mM MgCl2. The reactions were run at 85 °C for 1 min followed by 63 °C for 20 min. After cascade invasive reaction, 3 μl of each AuNPs (30 nM), NaCl (500 mM) and water were added in a volume of 30 μl. Hybridization was performed at 55 °C for 30 min. The results were observed by naked eyes directly or after briefly centrifuging the products. The positive reaction mixture kept red while the negative reaction mixture became colorless.
Sensitivity and specificity of mRT-LAMP-IRNH
Ten-fold serial dilutions of synthetic RNA transcripts of the three HA genes (avian influenza A/H5, A/H7 and 2009A/H1, ranging from 104 to100 RNA copies) were used to assess the analytical sensitivity of the mRT-LAMP-IRNH assay. The specificity of the assay was determined by analyzing the RNA extracts from various control viruses mentioned above. Briefly, RNA extracted from influenza viruses A/Nanjing/2/2013(H7N9), A/Jiangsu/1/2007(H5N1), A/Jiangsu/2/2009(H1N1), or each control virus was used as RT-LAMP template, respectively. The amplicons from each RT-LAMP reaction were further identified by cascade invasive reaction and gold nanoparticle hybridization as described before in three separate target-specific detection tubes.
Clinical specimen analysis
To investigate the feasibility of our methodology for clinical sample analysis, a total of 88 clinical specimens were collected and analyzed by mRT-LAMP-IRNH assay with the parallel analysis by our in-house real-time RT-PCR assays for influenza A/H5, A/H7 and 2009A/H1. The primers and probes used in our real-time RT-PCR assays for A/H5, A/H7 and 2009A/H1 were all recommended by WHO48,49,50, and all the three assays had been validated against viral culture and commercial real-time RT-PCR kits. The Real-time RT-PCR was performed using a SuperScript® III Platinum One-Step qRT-PCR System (Invitrogen) according to the instructions.
Additional Information
How to cite this article: Chi, Y. et al. Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Cascade Invasive Reaction and Nanoparticle Hybridization for Subtyping of Influenza A Virus. Sci. Rep. 7, 44924; doi: 10.1038/srep44924 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Simonsen, L. et al. Global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: a modeling study. PLoS Med 10, e1001558, doi: 10.1371/journal.pmed.1001558 (2013).
Medina, R. A. & Garcia-Sastre, A. Influenza A viruses: new research developments. Nat Rev Microbiol 9, 590–603, doi: 10.1038/nrmicro2613 (2011).
Fouchier, R. A. et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79, 2814–2822, doi: 10.1128/JVI.79.5.2814-2822.2005 (2005).
Harper, S. A. et al. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 53, 1–40 (2004).
Epperson, S. et al. Influenza activity - United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb Mortal Wkly Rep 63, 483–490 (2014).
Uyeki, T. M. Preventing and controlling influenza with available interventions. N Engl J Med 370, 789–791, doi: 10.1056/NEJMp1400034 (2014).
Hien, T. T., de Jong, M. & Farrar, J. Avian influenza–a challenge to global health care structures. N Engl J Med 351, 2363–2365, doi: 10.1056/NEJMp048267 (2004).
Beigel, J. H. et al. Avian influenza A (H5N1) infection in humans. N Engl J Med 353, 1374–1385, doi: 10.1056/NEJMra052211 (2005).
Yang, Z. F., Mok, C. K., Peiris, J. S. & Zhong, N. S. Human Infection with a Novel Avian Influenza A(H5N6) Virus. N Engl J Med 373, 487–489, doi: 10.1056/NEJMc1502983 (2015).
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368, 1888–1897, doi: 10.1056/NEJMoa1304459 (2013).
Ma, X. et al. A multiplex PCR assay for the detection of five influenza viruses using a dual priming oligonucleotide system. BMC Infect Dis 15, 93, doi: 10.1186/s12879-015-0818-y (2015).
Chaharaein, B., Omar, A. R., Aini, I., Yusoff, K. & Hassan, S. S. Detection of H5, H7 and H9 subtypes of avian influenza viruses by multiplex reverse transcription-polymerase chain reaction. Microbiol Res 164, 174–179, doi: 10.1016/j.micres.2007.01.001 (2009).
Suwannakarn, K. et al. Typing (A/B) and subtyping (H1/H3/H5) of influenza A viruses by multiplex real-time RT-PCR assays. J Virol Methods 152, 25–31, doi: 10.1016/j.jviromet.2008.06.002 (2008).
Choi, J. H. et al. Development and evaluation of multiplex real-time RT-PCR assays for seasonal, pandemic A/H1pdm09 and avian A/H5 influenza viruses detection. J Microbiol 51, 252–257, doi: 10.1007/s12275-013-2452-y (2013).
Munro, S. B., Kuypers, J. & Jerome, K. R. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J Clin Microbiol 51, 1124–1129, doi: 10.1128/JCM.03113-12 (2013).
Niemz, A., Ferguson, T. M. & Boyle, D. S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol 29, 240–250, doi: 10.1016/j.tibtech.2011.01.007 (2011).
Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, E63 (2000).
Parida, M., Sannarangaiah, S., Dash, P. K., Rao, P. V. & Morita, K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 18, 407–421, doi: 10.1002/rmv.593 (2008).
Hong, T. C. et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol 42, 1956–1961 (2004).
Imai, M. et al. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 24, 6679–6682, doi: 10.1016/j.vaccine.2006.05.046 (2006).
Curtis, K. A., Rudolph, D. L. & Owen, S. M. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP). J Virol Methods 151, 264–270, doi: 10.1016/j.jviromet.2008.04.011 (2008).
Wang, X. et al. Detection of enterovirus 71 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). J Virol Methods 179, 330–334, doi: 10.1016/j.jviromet.2011.11.019 (2012).
Ge, Y. et al. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS One 8, e69941, doi: 10.1371/journal.pone.0069941 (2013).
Liang, T. et al. A rapid assay for simultaneous detection of Spiroplasma eriocheiris and white spot syndrome virus in Procambarus clarkii by multiplex PCR. Lett Appl Microbiol 51, 532–538, doi: 10.1111/j.1472-765X.2010.02927.x (2010).
Liang, C. et al. Multiplex loop-mediated isothermal amplification detection by sequence-based barcodes coupled with nicking endonuclease-mediated pyrosequencing. Anal Chem 84, 3758–3763, doi: 10.1021/ac3003825 (2012).
Mahony, J. et al. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol 58, 127–131, doi: 10.1016/j.jcv.2013.06.006 (2013).
Tanner, N. A., Zhang, Y. & Evans, T. C. Jr. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques 53, 81–89, doi: 10.2144/0000113902 (2012).
Wang, Y. et al. Multiple Endonuclease Restriction Real-Time Loop-Mediated Isothermal Amplification: A Novel Analytically Rapid, Sensitive, Multiplex Loop-Mediated Isothermal Amplification Detection Technique. J Mol Diagn 17, 392–401, doi: 10.1016/j.jmoldx.2015.03.002 (2015).
Xu, W., Xue, X., Li, T., Zeng, H. & Liu, X. Ultrasensitive and selective colorimetric DNA detection by nicking endonuclease assisted nanoparticle amplification. Angew Chem Int Ed Engl 48, 6849–6852, doi: 10.1002/anie.200901772 (2009).
Xu, W. et al. Ultrasensitive colorimetric DNA detection using a combination of rolling circle amplification and nicking endonuclease-assisted nanoparticle amplification (NEANA). Small 8, 1846–1850, doi: 10.1002/smll.201200263 (2012).
Zou, B. et al. Sensitive and specific colorimetric DNA detection by invasive reaction coupled with nicking endonuclease-assisted nanoparticles amplification. Biosens Bioelectron 66, 50–54, doi: 10.1016/j.bios.2014.10.077 (2015).
Lyamichev, V. I. et al. Experimental and theoretical analysis of the invasive signal amplification reaction. Biochemistry 39, 9523–9532 (2000).
Hall, J. G. et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci USA 97, 8272–8277, doi: 10.1073/pnas.140225597 (2000).
Alexander, D. J. Avian influenza - diagnosis. Zoonoses Public Health 55, 16–23, doi: 10.1111/j.1863-2378.2007.01082.x (2008).
Mori, Y., Kitao, M., Tomita, N. & Notomi, T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods 59, 145–157, doi: 10.1016/j.jbbm.2003.12.005 (2004).
Nyan, D. C. & Swinson, K. L. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci Rep 5, 17925, doi: 10.1038/srep17925 (2015).
Tomita, N., Mori, Y., Kanda, H. & Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3, 877–882, doi: 10.1038/nprot.2008.57 (2008).
Tao, Z. Y. et al. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors 4, 115, doi: 10.1186/1756-3305-4-115 (2011).
Dou, M. et al. Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosens Bioelectron 87, 865–873, doi: 10.1016/j.bios.2016.09.033 (2016).
Osman, H. A. et al. Development and evaluation of loop-mediated isothermal amplification assay for detection of Crimean Congo hemorrhagic fever virus in Sudan. J Virol Methods 190, 4–10, doi: 10.1016/j.jviromet.2013.03.004 (2013).
Wang, C., Shen, X., Lu, J. & Zhang, L. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) system for rapid detection of HDV genotype 1. Lett Appl Microbiol 56, 229–235, doi: 10.1111/lam.12039 (2013).
Kaneko, H., Kawana, T., Fukushima, E. & Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods 70, 499–501, doi: 10.1016/j.jbbm.2006.08.008 (2007).
Nakauchi, M., Takayama, I., Takahashi, H., Tashiro, M. & Kageyama, T. Development of a reverse transcription loop-mediated isothermal amplification assay for the rapid diagnosis of avian influenza A (H7N9) virus infection. J Virol Methods 204, 101–104, doi: 10.1016/j.jviromet.2014.03.028 (2014).
Lee, M. S. et al. M-specific reverse transcription loop-mediated isothermal amplification for detection of pandemic (H1N1) 2009 virus. Eur J Clin Invest 41, 434–441, doi: 10.1111/j.1365-2362.2010.02427.x (2011).
Ma, X. J. et al. Visual detection of pandemic influenza A H1N1 Virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. J Virol Methods 167, 214–217, doi: 10.1016/j.jviromet.2010.03.027 (2010).
Hill, H. D. & Mirkin, C. A. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat Protoc 1, 324–336, doi: 10.1038/nprot.2006.51 (2006).
Song, Q. et al. Invader Assisted Enzyme-Linked Immunosorbent Assay for Colorimetric Detection of Disease Biomarkers Using Oligonucleotide Probe-Modified Gold Nanoparticles. J Biomed Nanotechnol 12, 831–839 (2016).
WHO. Real-time RT-PCR Protocol for the Detection of Avian Influenza A(H7N9) Virus. Available: http://www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf. Accessed 2016 October 10 (2013).
WHO. CDC protocol of realtime RTPCR for influenza A(H1N1). Available: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf. Accessed 2016 October 10 (2009).
WHO. Recommendations and laboratory procedures for detection of avian influenza A(H5N1) virus in specimens from suspected human cases. Available: http://www.who.int/influenza/resources/documents/RecAIlabtestsAug07.pdf. Accessed 2016 October 10 (2007).
Acknowledgements
The study was supported in part by the Important National Science & Technology Specific Projects during the twelve five-year plan period (2013ZX10004103-006), Jiangsu Province Science & Technology Demonstration Project for Emerging Infectious Diseases Control and Prevention (No. BE2015714), the National Natural Science Foundation of China (81501785, 81601732, 61401217, 81302466), the Natural Science Foundation of Jiangsu Province (BK20141030, BK20161583, BK20140900), and the “333” Projects of Jiangsu Province.
Author information
Authors and Affiliations
Contributions
C.S. and L.C. conceived and designed the experiments; Y.C., Y.G., K.Z., X.Q. and B.Z. performed the experiments; L.C., Y.C., Y.G. and B.L. analyzed the data; Y.C., Y.G. and Q.B. wrote the manuscript; Z.S., F.Z. and M.Z. reviewed manuscript. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chi, Y., Ge, Y., Zhao, K. et al. Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Cascade Invasive Reaction and Nanoparticle Hybridization for Subtyping of Influenza A Virus. Sci Rep 7, 44924 (2017). https://doi.org/10.1038/srep44924
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44924
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.