Abstract
Eukaryotic genomes are transcribed into numerous regulatory long non-coding RNAs (lncRNAs). Compared to mRNAs, lncRNAs display higher developmental stage-, tissue-, and cell-subtype-specificity of expression, and are generally less abundant in a population of cells. Despite the progress in single-cell-focused research, the origins of low population-level expression of lncRNAs in homogeneous populations of cells are poorly understood. Here, we identify HIPSTR (Heterogeneously expressed from the Intronic Plus Strand of the TFAP2A-locus RNA), a novel lncRNA gene in the developmentally regulated TFAP2A locus. HIPSTR has evolutionarily conserved expression patterns, its promoter is most active in undifferentiated cells, and depletion of HIPSTR in HEK293 and in pluripotent H1BP cells predominantly affects the genes involved in early organismal development and cell differentiation. Most importantly, we find that HIPSTR is specifically induced and heterogeneously expressed in the 8-cell-stage human embryos during the major wave of embryonic genome activation. We systematically explore the phenomenon of cell-to-cell variation of gene expression and link it to low population-level expression of lncRNAs, showing that, similar to HIPSTR, the expression of thousands of lncRNAs is more highly heterogeneous than the expression of mRNAs in the individual, otherwise indistinguishable cells of totipotent human embryos, primordial germ cells, and stable cell lines.
Similar content being viewed by others
Introduction
Eukaryotic genomes are pervasively transcribed1,2,3, producing thousands of uncharacterized transcripts, the majority of which are classified as long non-coding RNAs (lncRNAs) (ref. 4). LncRNAs are simply defined as long (>200 nt) non-protein coding transcripts, and as such they represent a very broad, widely uncharacterized group that includes non-functional transcripts resulting from transcriptional noise (random transcription initiation by RNA Pol II throughout the genome), and lncRNAs exerting their function either passively through the act of their transcription or actively in cis and in trans5. When compared to mRNAs, lncRNAs are expressed at lower levels with considerably higher tissue-specificity6, developmental stage-specificity7, and in heterogeneous tissues – also cell-subtype specificity8. It has been proposed that low expression levels of lncRNAs originate from high cell-to-cell variation in expression of lncRNA genes9. Nonetheless, arguing with this hypothesis and findings from mouse bone-marrow-derived dendritic cells10, a recent study exploited single-cell RNA-FISH and demonstrated that cell-to-cell variation of lncRNAs expression is similar to that of mRNAs11. Overall, the relationship of the low population-level expression of lncRNAs and cell-to-cell variation of gene expression remains largely unexplored.
Although, antisense transcription has been proposed to occur in 74% of human gene loci12, antisense lncRNAs remain the least studied group of all lncRNAs. Five prime regions of antisense lncRNA genes coincide with the presence of promoter-associated chromatin marks, CpG islands, and RNA Pol II binding13,14, and 28% of antisense transcripts were detected in the absence of expression of their overlapping genes15, further supporting the independence of these transcription units. Our previous work demonstrates that such antisense transcription units frequently produce monoexonic lncRNAs16. The most highly expressed antisense lncRNAs are transcribed antisense to genes encoding transcription regulators12. The importance of antisense lncRNAs17 is illustrated by a particular example of ANRASSF1 (ref. 18) oncogenic lncRNA, and by differential expression of multiple antisense lncRNAs in pancreatic cancer13, and renal cell carcinoma14, where the expression of antisense lncRNAs is correlated with expression of their sense counterparts12,14,16. The widely accepted assumption that a large portion of antisense lncRNAs regulates their overlapping genes19 might nonetheless be a poor predictor of function for any yet uncharacterized antisense lncRNA.
In this study, we identified a novel lncRNA, which we named HIPSTR, that is expressed from the opposite strand of TFAP2A, the gene encoding a transcription factor (TF) involved in tumorigenesis20, and important for neural crest21,22,23,24 and trophectoderm25,26 development. We found that HIPSTR has conserved expression patterns between human and mouse, and that its promoter demarcation is conserved in the amniotes. Unlike previously characterized antisense lncRNAs, we found that HIPSTR expression levels do not correlate with the expression of its overlapping TFAP2A gene in cell lines, tissues, and developmental models. Silencing of HIPSTR led to differential expression of a group of genes involved in development and differentiation. Consistently, we show that HIPSTR is activated independently from TFAP2A during early development, where it has heterogeneous expression, being expressed in only a subset of cells within totipotent human embryos. We further explore the phenomenon of heterogeneous expression and demonstrate that lncRNAs in totipotent human embryos, human embryonic stem cells (hESCs), human primordial germ cells (hPGCs), and myelogenous leukemia cells (K562) have significantly higher cell-to-cell variation in expression than mRNAs.
Results
HIPSTR is a bona fide antisense lncRNA with evolutionarily conserved expression patterns
Aiming at the identification of novel antisense lncRNAs possibly associated with prostate cancer, we obtained strand-specific deep RNA-seq data from LNCaP prostate cancer cell line and searched for antisense transcription events in loci encoding TFs. TFAP2A encodes a TF known to be involved in various cancers (reviewed in ref. 20), including prostate cancer27,28, where TFAP2A is downregulated, and its promoter is hypermethylated in LNCaP and DU145 model cell lines, as well as in clinical samples27. We focused on a putative monoexonic antisense lncRNA gene located between exons 2 and 5 of TFAP2A on the opposite genomic strand (Fig. 1A). We later named this lncRNA gene HIPSTR (Heterogeneously expressed from the Intronic Plus Strand of the TFAP2A-locus RNA).
HIPSTR is a bona fide lncRNA.
(A) Genomic position of human HIPSTR relative to the TFAP2A locus genes. The predicted HIPSTR polyadenylation signal is marked with a red “X” sign; genomic coordinates of the region shown are hg19 chr6:10396400–10420700. (B) RNA Pol II inhibition by α-amanitin in HeLa cells decreases HIPSTR levels; known RNA Pol II transcripts (ACTB, MYC) and RNA Pol III transcripts (pre-tRNATyr, 7SK) served as controls. (C) 5′-cap structure removal by co-treatment of HeLa cells total RNA with Terminator 5′-phosphate-dependent exonuclease (Ter) and tobacco acid pyrophosphatase (TAP) reduces levels of HIPSTR; capped TUBA1C and uncapped SNORD15A transcripts served as controls. (D) HeLa cells fractionation shows nuclear enrichment of HIPSTR; nuclear enrichment of TFAP2A and TFAP2A-AS1 is comparable with that of ACTB; TFAP2A pre-mRNA, MALAT1 lncRNA, and 45S rRNA served as nuclear fraction controls; 18S rRNA served as cytoplasmic fraction control. The same RNA samples were used as in ref. 69, and data shown on (B–D) for control transcripts are the same as presented on Fig. 3A,B,D in ref. 69. (E) HIPSTR expression cannot be associated with tumor or non-tumor phenotype, as measured in human tumor (solid bars) and non-tumor (hatched bars) cell lines; expression in non-tumor HEK293 cell line (hatched green bar) is shown for comparison. (F) HIPSTR expression does not correlate with TFAP2A levels in the human cell lines shown on (E). (G) HIPSTR expression does not correlate with TFAP2A levels in the ENCODE Project29 RNA-seq data from human cell lines (A549, GM12878, H1 hESCs, HeLa-S3, HepG2, HMEC, HSMM, HUVEC, K562, MCF7, NHEK). (H) HIPSTR expression does not correlate with TFAP2A levels in the human tissues shown on (J) (see below). (I) Mouse Hipstr (mm9 chr13:40818458–40821725) ortholog expression across a panel of mouse tissue RNA samples from the Mouse ENCODE Project70 RNA-seq data. (J) HIPSTR expression across a panel of human tissue RNA samples. Data shown on (B–F,H,J) are RT-qPCR read-outs of three independent experiments, error bars represent SD; data on (I,G) is our re-analysis of public RNA-seq; N/D – not detected.
We combined our results of RACE PCR (Fig. S1A), with our and public RNA-seq data to obtain the full-length sequence of the unspliced polyadenylated HIPSTR lncRNA (3427 nt, chr6:10404735–10408161 in human genome assembly hg19; Fig. 1A). Analysis of ENCODE Project data29 showed that HIPSTR has an alternative TSS in HeLa-S3 cells, located more than 600 bp upstream of the HIPSTR TSS in LNCaP or K562 cells (Fig. S1A). It remains to be investigated whether this alternative HIPSTR isoform is functionally different from the HIPSTR isoform described in this study. It is also evident from RNA-seq data that HIPSTR transcripts are unspliced (Fig. 1A, S1A). HIPSTR TSS is located within an 818-bp-long CpG island (Fig. 1A) and overlaps RNA Pol II ChIP-seq peaks from ENCODE Project data29 (Fig. S1B). We confirmed that HIPSTR is transcribed by RNA Pol II (Fig. 1B), and has a 5′-cap structure (Fig. 1C).
We next examined HIPSTR coding potential. First, we observed a strong nuclear enrichment of HIPSTR transcript (~33.5-fold, Fig. 1D), similar to some previously described regulatory nuclear lncRNAs (see Table 1 in ref. 30). In the nucleus, HIPSTR is associated with chromatin through the first 1000 nt of its sequence (Fig. S1C), although it is not possible to determine whether HIPSTR lncRNA remains associated with the chromatin at the same locus where it is produced. Both CPC (ref. 31) and CPAT (ref. 32) coding potential evaluation tools classified HIPSTR as non-coding. None of the potential ORFs within HIPSTR sequence showed any similarity to known proteins in a blastx search. There was no evidence of significant ribosome association with the HIPSTR sequence in the ribosome profiling data from ref. 33 (Fig. S1D). Finally, in silico analysis demonstrated that the longest potential ORF in the HIPSTR sequence (345 nt) can be expected to occur by chance in a 3427 nt-long transcript (Fig. S1E). Altogether, these data argue that HIPSTR is a bona fide lncRNA.
Considering the proposed roles for antisense RNAs in cancer13,14, we hypothesized that HIPSTR may be differentially expressed in tumor and non-tumor cell lines. We found that HIPSTR expression was not associated with tumor or non-tumor phenotype in prostate, kidney, breast, liver or endometrial cell lines (Fig. 1E). Moreover, HIPSTR expression did not correlate with its overlapping gene (TFAP2A) across the cell lines tested (Fig. 1F). The latter observation was further supported by analysis of HIPSTR and TFAP2A expression in ENCODE Project RNA-seq data sets29 (Fig. 1G), and in a panel of human tissue RNA samples (Fig. 1H). Consistent with previous reports6,34 for lncRNAs, we found that HIPSTR population-level expression was low and exceeded the value of 1 FPKM only in two (HeLa-S3 and K562) out of eleven ENCODE cell lines29 analyzed here (Fig. 1G).
We successfully detected HIPSTR transcription with RT-qPCR in a panel of mouse tissue RNA samples (Fig. S1F). Finally, we found that HIPSTR has an evolutionarily conserved tissue-specific expression pattern, and that it is predominantly expressed in testis and placenta of mouse (Mouse ENCODE Project RNA-seq data29 analyzed on Fig. 1I) and of human (Fig. 1J).
HIPSTR promoter demarcation is conserved in the amniotes
The highest level of turnover among all classes of functional elements identified by the ENCODE Project35, and the lack of known orthologs in other species are common features of lncRNAs (reviewed in refs 36, 37). Only 19% of lncRNA families expressed in three or more tetrapod species studied by Necsulea et al.38 did originate more than 90 million years ago (Ma). In addition, only 21% of lncRNA loci that are present in human, chimpanzee, and macaque have an orthologous lncRNA outside of primates38. Interestingly, human lncRNAs transcribed from canonical RNA Pol II promoters emit strong and consistent signal of purifying selection, as opposed to lncRNAs transcribed from enhancers39. Notably, a characteristic promoter-associated H3K4me3 mark40,41 can be present on active and silent promoters40,41. We found that HIPSTR TSS was indeed demarcated by H3K4me3 (Fig. S2A) in K562 and NT2/D1 cells, which have high and undetectable levels of HIPSTR, respectively.
We first questioned the ability of DNA sequences surrounding HIPSTR TSS and occupied by H3K4me3 mark in K562 and NT2/D1 (ref. 29) to drive reporter gene transcription in four human cell lines (HeLa, HEK293, HepG2 and NT2/D1). We cloned sequences surrounding HIPSTR TSS (constructs pGL3-P1 to -P7, Fig. S2A) upstream of the firefly luciferase gene, and compared the luminescence signal produced by cells transfected with these reporters. We tested seven sequences, and they produced ~3- to ~903-fold stronger luminescence signal than negative control plasmid (pGL3-Basic) in the cell lines tested (Fig. S2B). Although we found that the endogenous HIPSTR gene is not expressed in NT2/D1 embryonal carcinoma cells, in this pluripotent cell line two HIPSTR promoter-luciferase constructs (pGL3-P1 and pGL3-P3) produced ~35–50-times stronger luminescence signal than did positive control construct (pGL3-SV40) (Fig. S2B). These data implies that specifically in pluripotent cells a strong positive regulator is present that would be able to drive transcription from HIPSTR promoter located on a plasmid. This is not the case for the endogenous HIPSTR promoter in NT2/D1 cells, likely due to the presence of H3K27me3 repressive chromatin mark in the TFAP2A locus (Fig. S2A). The latter explanation is further supported by the observation that the initial strong luminescence signal from HIPSTR promoter-luciferase constructs in NT2/D1 cells decreases as they lose pluripotency along the course of ATRA treatment (Fig. S2C).
Since HIPSTR expression patterns are conserved between human and mouse (Fig. 1I,J), we asked whether other mammalian species also have the HIPSTR gene. Since no deep strand-specific RNA-seq data sets are available for placenta and testis for organisms other than human and mouse, we hypothesized that the presence of a H3K4me3 mark may help to indirectly estimate the degree of HIPSTR promoter conservation and hence – of HIPSTR transcription unit itself.
We analyzed public ChIP-seq data for various organisms42,43,44,45, and found H3K4me3 ChIP-seq peaks around HIPSTR TSS orthologous region in the samples of all 10 mammals tested, and of rooster, but not in any of the frog or zebrafish embryos (Fig. 2A). These results suggest that functional HIPSTR promoter demarcation existed approximately 325 Ma in a common ancestor of human and chicken36, and that therefore other amniotes likely have the HIPSTR gene.
TFAP2A can regulate HIPSTR promoter, but HIPSTR and TFAP2A are not consistently co-induced in in vitro developmental models.
(A) Re-analyses of H3K4me3 ChIP-seq data from refs 42, 43, 44, 45 reveal conserved HIPSTR promoter demarcation across the genomes of 10 mammalian species (see Methods) and of chicken, and absence of H3K4me3 mark around HIPSTR TSS orthologous region in frog and zebrafish. The maximal value on the y-axis scale corresponds to the highest H3K4me3 peak across the entire TFAP2A locus for each species. (B) Positions of the mapped H3K4me3 and TFAP2A ChIP-seq reads from ref. 29 (HeLa-S3 cells) and ref. 24 (three human NCC and two chimp NCC lines), and positions of the DNA sequences used for HIPSTR promoter-reporter assays (pGL3-P1 to -P7) relative to the TFAP2A locus genes. (C) Luciferase reporter assays in HEK293 cells upon TFAP2A isoform 1a overexpression. DNA sequences surrounding HIPSTR TSS (see above) were cloned upstream of the firefly luciferase gene, and co-transfected with plasmid overexpressing TFAP2A isoform 1a or with empty vector; pGL3-Basic served as negative control (no promoter upstream of the firefly luciferase); pGL3-SV40 served as positive control (SV40 promoter upstream of the firefly luciferase); 3xAP2bluc served as positive control for transactivation by TFAP2A isoform 1a. (D) Overexpression of TFAP2A isoforms 1a (dark red), 1b (red), or 1c (pink) upregulates endogenous HIPSTR in HEK293 cells, as measured with RT-qPCR. (E–G) HIPSTR is moderately co-upregulated with TFAP2A in in vitro derived human NCCs (E), weakly co-upregulated with TFAP2A in in vitro derived human TBCs (F), and not co-upregulated with TFAP2A in NT2/D1 cells treated with ATRA, where HIPSTR remains undetectable (G), as measured with RT-qPCR. Upregulation of TFAP2A gene itself (NCCs marker), of CGB (human TBCs marker), or HOXB5 gene (induced by ATRA treatment in NT2/D1 cells)47 served as positive controls in the corresponding experiments shown on (E–G). Experiments presented on (C–G) were performed in triplicate, and error bars represent SD. For experiments on (C,D) the asterisks indicate statistical significance of the observed changes calculated with two-tailed t-test, equal variance (p-value < 0.01).
HIPSTR promoter can be stimulated by TFAP2A, but HIPSTR and TFAP2A are not consistently co-induced in developmental models in vitro
Increased activity of HIPSTR promoter in pluripotent cells suggests that HIPSTR may be involved in early embryonic development. Moreover, HIPSTR overlaps the developmentally-regulated TFAP2A gene that is induced and plays important roles in neural crest cells21,22,23,24 (NCCs), and in trophoblast cells25,26,46 (TBCs). Finally, TFAP2A expression can be transiently induced in human embryonal carcinoma NT2/D1 cells grown in the presence of ATRA (ref. 47). Interestingly, we found that TFAP2A ChIP-seq peaks were mapped to sequences upstream and downstream of HIPSTR TSS in HeLa-S3 cells (data from ref. 29), in human NCCs (data from ref. 23), and in the corresponding orthologous regions in chimp NCCs (data from refs 23, 24) (Fig. 2B). To assess the importance of TFAP2A in the regulation of HIPSTR promoter, we first overexpressed TFAP2A isoform 1a, and observed a significant increase in the luminescence signal from HIPSTR promoter-luciferase constructs in HEK293 (Fig. 2C), but not in HepG2 cells (Fig. S2D), while the activity of the TFAP2A-specific reporter (3xAP2bluc) increased in both experiments. To date, three TFAP2A isoforms with different functions were described48, and we next overexpressed each isoform in HEK293 and HepG2 cells (Fig. S2E), and measured expression of the endogenous HIPSTR. When overexpressed, each TFAP2A isoform upregulated endogenous HIPSTR in HEK293 cells (Fig. 2D), but was unable to initiate HIPSTR expression in HepG2 cells that lack endogenous HIPSTR expression (Fig. S2F). Aside from TFAP2A, dozens of TFs bind HIPSTR promoter, when assayed in different cell lines, as seen from ENCODE Project29 ChIP-seq data. Future studies should evaluate their relevance for HIPSTR promoter regulation in different cells.
Since HIPSTR gene is completely overlapped by developmentally regulated TFAP2A gene, and can be regulated by the protein product of the latter, we next hypothesized that both genes could be simultaneously induced during development. We induced TFAP2A expression in vitro in the three model systems mentioned above (hNCCs, hTBCs, and ATRA-treated NT2/D1 cells), and monitored HIPSTR expression levels. We confirmed a strong induction of TFAP2A transcription, and observed a simultaneous upregulation of divergently transcribed TFAP2A-AS1 lncRNA in all three systems (Fig. 2E–G). However, >100-fold upregulation of TFAP2A was accompanied by only ~9.4-fold induction of HIPSTR in hNCCs (Fig. 2E) and by ~1.8-fold induction of HIPSTR in hTBCs (Fig. 2F). Moreover, ATRA treatment of NT2/D1 cells failed to induce HIPSTR expression (Fig. 2G). These results demonstrate the lack of consistent and robust co-induction of HIPSTR and TFAP2A in developmental models.
HIPSTR silencing in HEK293 and H1BP cells affects development-associated genes
HIPSTR levels do not correlate with the expression of TFAP2A. We then reasoned that chromatin-associated HIPSTR lncRNA might regulate other genes elsewhere in the genome in trans. Consistent with a relatively short half-life of this lncRNA (38 min, Fig. S3A), efficient HIPSTR silencing in HEK293 cells with a pool of targeting ASOs was achieved as early as 6 h after transfection (~71%, Fig. S3B), and the highest efficiency was reached 24 h post-transfection (~89%, Fig. S3B). HIPSTR silencing in HEK293 cells with each of the two targeting ASOs separately (ASO #1 and ASO #2; Fig. 3A) did not affect the overall levels of TFAP2A expression (Fig. 3A), but instead it significantly altered TFAP2A-AS1 expression (Fig. 3A; see further details below). Neither the mRNA levels of specific TFAP2A isoforms, nor the levels of TFAP2A pre-mRNA were affected (Fig. 3B). HIPSTR knockdown resulted in genome-wide differential expression of 380 annotated genes (439 probes) located outside of the TFAP2A locus (Fig. 3C, Table S1), of which 377 (~99.2%) were upregulated. These results suggest a repressive function for HIPSTR in HEK293 cells. Accordingly, transient overexpression of HIPSTR in HEK293 cells (Fig. S3C) resulted in downregulation (Fig. S3D) of eight out of the twelve selected genes that were upregulated in HIPSTR knockdown experiments (Table S1). At the same time, Gene Ontology (GO) analysis of the genes upregulated by HIPSTR knockdown revealed their enrichment in “Developmental Process” and “Cell Differentiation” categories (Fig. S3E). A group of genes upregulated by HIPSTR knockdown in HEK293 cells was also upregulated by HIPSTR silencing in LNCaP cells (Fig. S3F), further validating these results.
Developmental genes are affected by HIPSTR knockdown in HEK293 and H1BP cells.
(A) Effect of HIPSTR knockdown on the expression of TFAP2A locus genes in HEK293 cells. (B) HIPSTR knockdown does not significantly affect (p-value < 0.05, fold-change >2) the abundance of TFAP2A isoforms or pre-mRNA. (C) Heat map showing that HIPSTR knockdown in HEK293 cells leads to a significant upregulation of 377 annotated genes outside of TFAP2A locus (1% FDR, fold-change >2, also see Table S1). (D) Efficiency of HIPSTR knockdown in H1BP cells. (E) Overlap between genes differentially expressed upon HIPSTR silencing in HEK293 and H1BP cells (also see Table S2). (F) Validation of a group of genes, whose expression is significantly up- and downregulated by HIPSTR knockdown in HEK293 (top panel) and H1BP cells (bottom panel), correspondingly. (G) Heat map demonstrating that HIPSTR knockdown in H1BP cells leads to significant upregulation of 572 and downregulation of 777 genes (1% FDR, fold-change >2, also see Table S3). Data shown on (A,B,D,F) are RT-qPCR read-outs of three independent experiments, error bars represent SD; N/D – not detected; the asterisks indicate statistical significance of the expression differences (fold-change >2) calculated with two-tailed t-test, equal variance (p-value < 0.05).
Finally, we noted that in HEK293 cells, HIPSTR knockdown also did upregulate TFAP2A-AS1 lncRNA (Fig. 3A). In turn, TFAP2A-AS1 silencing with two different ASOs (ASO #3 and ASO #4; Fig. S3G) did upregulate HIPSTR (Fig. S3G) and several genes that were upregulated by HIPSTR knockdown (Fig. S3H, S3I). The upregulation of these genes (except for ZSCAN10) was inversely proportional to the extent of upregulation of HIPSTR upon TFAP2A-AS1 knockdown with these ASOs (Fig. S3G, S3H), suggesting a partially overlapping function for these lncRNAs in HEK293 cells. Finally, TFAP2A protein levels in HEK293 cells were not affected by HIPSTR knockdown (Fig. S4A) and overexpression experiments (Fig. S4B).
We had established that HIPSTR promoter is most active in undifferentiated cells, but we could not induce HIPSTR expression with ATRA treatment of pluripotent carcinoma NT2/D1 cells. Conveniently, pluripotent H1BP cells have a normal karyotype, express higher levels of HIPSTR than H1 hESCs from which they were derived (Fig. S4C), and have been proposed to be analogous to the outer cells of the 16-cell morula49. We silenced HIPSTR in H1BP cells with three ASOs (ASO #1 and ASO #2 along with an additional ASO #0, Fig. 3D). HIPSTR knockdown did not change the morphology of H1BP cells or expression of the core pluripotency network genes (Table S3). TFAP2A and TFAP2A-AS1 remained undetectable (Fig. 3D), pointing at an independence of HIPSTR from TFAP2A-AS1, even though they may co-regulate a set of genes in HEK293 cells. Genome-wide, 49 annotated genes (53 probes) upregulated upon HIPSTR knockdown in HEK293 cells showed downregulation after HIPSTR depletion in H1BP cells (Fig. 3E, Table S2), and we validated with RT-qPCR such an opposite differential expression pattern for a group of these genes after HIPSTR knockdown in HEK293 (Fig. 3F, top) and H1BP cells (Fig. 3F, bottom). Overall, HIPSTR silencing in H1BP cells resulted in 1349 significantly differentially expressed annotated genes (Fig. 3G, Table S3), of which 777 genes (985 probes; ~62.2%; Fig. 3G) were downregulated and showed enrichment in development- and metabolism-related GO categories (Fig. S4D). The remaining 572 genes (598 probes; ~37.8%; Fig. 3G) were upregulated and enriched for skin-, placenta-, lung-, and brain-specific expression (Fig. S4E). These results suggest that in the context of a pluripotent cell (H1BP cells), HIPSTR is capable of both activating and repressing its target genes, whereas in a cell lacking pluripotency network associated factors (HEK293, LNCaP cells) HIPSTR acts solely as a repressor.
HIPSTR expression in the early human embryo is restricted to a subset of cells
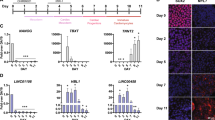
We next addressed a possibility that HIPSTR induction occurs prior to and independent from the activation of TFAP2A in trophectoderm and/or neural crest during the course of embryonic development. In the past few years, several studies reported successful transcriptome sequencing of individual blastomeres of early human and mouse embryos7,50,51,52. We thus mapped (Fig. 4A) public RNA-seq data from two data sets7,50, and quantified HIPSTR expression in these data (Fig. 4C,D). As these RNA-seq data sets are not strand-specific, we present HIPSTR expression as “underestimated” and “overestimated” FPKM values, by accordingly excluding or including the reads mapping to exons of TFAP2A that overlap HIPSTR. We found that HIPSTR, and not TFAP2A or TFAP2A-AS1 (Fig. 4C–F), is specifically upregulated in 8-cell and likely morula stage human embryos. We confirmed the presence of HIPSTR expression in 2–3 days-old human embryos (in one cell from a 4-cell stage embryo, and in eight cells from five separate 8-cell stage embryos) in the strand-specific single-cell-tagged reverse-transcription STRT-seq libraries from ref. 51 (Fig. 4B). These data indicate that HIPSTR gene is activated shortly after a major wave of human embryonic genome activation7 (EGA). It is evident from our analysis of public RNA-seq data from ref. 52 that expression of mouse Hipstr occurs in 2-cell embryos, soon after mouse EGA (Fig. S5B–S5D). Nonetheless, these latter observations are in conflict with mouse single-cell RNA-seq data from ref. 50, where we saw no evidence of expression in the HIPSTR orthologous region at all stages, including 2-cell stage (not shown). Hence, our results suggest that HIPSTR likely functions during the major wave of EGA in human embryos, but whether this is the case for mouse embryonic development remains an open question. Most intriguingly, we found that during the major wave of EGA within the 8-cell and morula stage human embryos (Fig. 4G,H), and in a population of K562 cells (Fig. S5A), HIPSTR expression is restricted to only a subset of cells.
HIPSTR is expressed by a subset of cells in early human embryos.
(A) Mapping of RNA-seq reads from ref. 7 illustrates specific expression of HIPSTR, and not TFAP2A or TFAP2A-AS1, in 8-cell and morula-stage human embryos. (B) Mapping of the 5′-ends of transcripts with strand-specific STRT-seq data from ref. 51 shows specific expression of HIPSTR in one cell (4b2) from a 4-cell human embryo, and in eight cells (8c6 through 8i6) originating from five different 8-cell human embryos; cell names are as in ref. 51. (C,D) Re-analyses of aggregate RNA-seq data for each developmental stage from ref. 7 on (C), and from ref. 50 on (D) confirms that HIPSTR induction in early embryos is independent from TFAP2A and TFAP2A-AS1 genes. (E,F) TFAP2A pre-mRNA is not detectable in human oocytes and early embryos; re-analyses of aggregate RNA-seq data from ref. 7 on (E), and from ref. 50 on (F). (G,H) HIPSTR expression is restricted to a subset of cells in early human embryos, as inferred from RNA-seq data for 8-cell- and morula-stage embryos from ref. 7 in (G), or RNA-seq data for 8-cell-stage embryos from ref. 50 in (H); plotted are overestimated FPKM values for HIPSTR expression (see text).
Cell-to-cell variability in expression of lncRNAs is higher than that of mRNAs
In a recent work, Cabili et al. used single-molecule RNA-FISH approach, and concluded that no difference exists in cell-to-cell variability in expression of mRNAs and lncRNAs11. This argues against a hypothesis that lncRNAs with low population-level abundance are instead expressed at high levels by a subset of cells within that population9. In agreement with the latter hypothesis are expression patterns of HIPSTR in early embryos and K562 cells, and of several mouse lncRNAs in bone-marrow-derived dendritic cells10.
To resolve this discrepancy between single-molecule RNA-FISH results and observations from single-cell RNA-seq data, we next systematically explored patterns of cell-to-cell expression variability of lncRNAs and mRNAs in human cells. For this, we used five single-cell RNA-seq data sets – from human totipotent blastomeres (36 cells; ref. 7), from pluripotent hESCs (32 cells; ref. 7), from K562 cells (96 cells; ref. 53), and from hPGCs of 7 weeks-old (7W; 39 cells; ref. 54) and of 19 weeks-old (19W; 57 cells, ref. 54) male embryos. We considered all expressed genes, defined here as those having max expression >3 FPKM (30-fold more stringent threshold than in refs 7, 54; see Methods) in at least one individual cell of a given data set, and compared the coefficient of variation of gene expression across the cells between lncRNAs and mRNAs. For genes with max expression within 3–30 FPKM, we saw a greater difference between non-coding and protein-coding transcripts than for those with max expression >30 FPKM (Fig. 5A–E). For the former group, the distribution of the numbers of cells was a mixture distribution. We fitted this mixture distribution with a finite mixture model with two populations, and used this model to classify lncRNAs and mRNAs as having high, low or uncertain heterogeneity of expression (Fig. 5F–J). For lncRNAs of this group (max expression 3–30 FPKM), only a small fraction showed low or uncertain (posterior probability <0.99) heterogeneity of expression −6.5%, 7.0%, 4.2%, 4.8%, and 2.3% in human totipotent blastomeres (Fig. 5F), hESCs (Fig. 5G), K562 cells (Fig. 5H), 7W hPGCs (Fig. 5I), and 19W hPGCs (Fig. 5J), respectively (Table S9). For example, in hESCs the known pluripotency regulator TUNAR (ref. 55) was assigned low heterogeneity flag in our analysis (Table S2). At the same time, HIPSTR was classified as a transcript with high heterogeneity of expression in 8-cell and morula-stage human embryos, and in K562 cells (Tables S4, S6), as expected. Remarkable heterogeneity of expression of lncRNAs was in a stark contrast to the much lower heterogeneity of expression of mRNAs with comparable expression levels (3–30 FPKM), of which 40%, 43%, 19%, 27%, and 20% were associated with low or uncertain heterogeneity of expression in human totipotent blastomeres, hESCs, K562 cells, 7W hPGCs, and 19W hPGCs, respectively (Fig. 5F–J; Table S9). Overall, lncRNAs analyzed here (max expression 3–30 FPKM) and assigned the high heterogeneity flag (H) constituted on average 74% of all expressed lncRNAs (>3 FPKM), while for mRNAs this fraction was only 35% (Table S8).
LncRNAs show higher heterogeneity of expression than mRNAs.
(A–E) LncRNAs have higher cell-to-cell variation in expression than mRNAs. Coefficient of variation (CV) across all cells of a given single-cell RNA-seq data set was calculated for each expressed gene (>3 FPKM), and shown are box plots of CV values for highly expressed (>30 FPKM) mRNAs (dark orange) and lncRNAs (dark grey), and for moderately expressed (3–30 FPKM) mRNAs (light orange) and lncRNAs (light grey). Box shows the first and third interquartile range (IQR), the line inside the box shows the median, and whiskers encompass the CV values within 1.5 IQR below and above the first and third quartiles, respectively. Points outside the whiskers are CV outliers. All possible pairwise comparisons result in statistically significant differences, Welch’s t-test (p-value < 0.001). (F–J) Higher fraction of lncRNAs is classified as highly heterogeneously expressed, as compared to mRNAs. Plotted are density distributions of numbers of expressing cells calculated for lncRNAs (black dashed line), mRNAs (red dashed line), lncRNAs and mRNAs together (grey bars), and for modeled populations of genes with high (solid light blue line) or low (solid dark blue line) heterogeneity of expression. Pie charts demonstrate fractions of lncRNAs and mRNAs associated with the population of genes with high (light blue), low (dark blue) or uncertain (grey) heterogeneity of expression. Genes used for this analysis had expression >3 FPKM in at least one cell, and <30 FPKM in all cells of the corresponding data sets. Genes that contributed to the plots and pie charts on (F–J) were classified as belonging to either of the modeled populations of genes (with high or low expression heterogeneity) with a posterior probability >0.99, or were assigned the “uncertain heterogeneity” classification otherwise (posterior probability ≤0.99) (Tables S4,S5,S6,S7,S8). Single-cell RNA-seq data sets re-analyzed here were from: ref. 7 (8-cell and morula stage embryos, hESCs), ref. 53 (K562 cells), and ref. 54 (7W hPGCs and 19 W hPGCs). Number of individual cells used for each analysis is in parentheses in each panel heading.
Discussion
In the present work, we searched for novel antisense lncRNAs in the loci encoding TFs, and identified HIPSTR gene (Heterogeneously expressed from the Intronic Plus Strand of the TFAP2A-locus RNA) that is located on the opposite strand of TFAP2A gene. HIPSTR is transcribed by RNA Pol II into a capped, monoexonic, nuclear-enriched, chromatin-associated antisense lncRNA (Fig. 1A–D, S1A–S1D). HIPSTR is a bona fide antisense lncRNA; it is not associated with ribosomes and does not possess ORFs that could potentially encode any known polypeptide.
Many antisense transcripts were shown to regulate their overlapping or divergently transcribed genes (reviewed in ref. 56). We found that HIPSTR is not consistently co-induced with its overlapping and developmentally-regulated TFAP2A gene in in vitro developmental models, and that HIPSTR levels do not correlate with the expression of TFAP2A in cell lines and tissues. Accordingly, HIPSTR expression perturbations in HEK293 and H1BP cells did not affect overall levels of TFAP2A mRNA (Fig. 3A,D), pre-mRNA (Fig. 3B), or TFAP2A protein levels (Fig. S4A). Genome-wide, HIPSTR likely acts context-dependently, as its knockdown upregulated a group of development-related genes (Fig. 3F, top) in non-pluripotent HEK293 cells, while in pluripotent H1BP cells these genes were downregulated by HIPSTR silencing (Fig. 3F, bottom).
Since HIPSTR is capable of regulating developmental genes in different systems (Fig. S3F, S4D), and does not correlate with TFAP2A, we have searched public data for an evidence of TFAP2A-independent activation of HIPSTR during early development. We found that HIPSTR expression is indeed induced independently of TFAP2A specifically in 8-cell embryos, during the major wave of human EGA (Fig. 4A–D). Whether conservation of HIPSTR expression pattern (Fig. 1A,J) extends to the major wave of mouse EGA (2-cell stage) remains to be established, as existing RNA-seq data for early mouse embryos are inconsistent with respect to Hipstr expression (Fig. S5B–S5D). Tfap2a-null mice die perinatally with severe congenital defects21,22. Most interestingly, Tfap2a−/− mice generated to date were obtained by targeting exons located upstream of the Hipstr gene and its promoter region. Should mouse Hipstr be induced in 2-cell embryos (and thus–prior to Tfap2a induction in trophectoderm or neural crest), genetic knockout studies would provide the ultimate evidence for the functional importance of HIPSTR during early embryonic development and the necessary support for mechanistic studies of HIPSTR function.
LncRNAs were proposed to function as modular scaffolds for chromatin modifying enzymes and TFs (ref. 57). Lower population-level expression of lncRNAs, as compared to mRNAs (refs 6, 34), might represent a serious obstacle for identification of partner proteins in RNA-Immunoprecipitation and endogenous RNA-pulldown assays, possibly resulting in false-negative results. We explore the origin of low population-levels of expression and show that lncRNAs are more heterogeneously expressed than mRNAs in individual, seemingly identical cells in vitro (Fig. 5A–C, Tables S4,S5,S6) and in vivo (Fig. 5D,E, Tables S7, S8). For example, in the K562 single-cell RNA-seq data set53, HIPSTR expression was completely absent (0 FPKM) from 73 individual cells, but reached 24.5 FPKM in 1 out of the 96 cells in that data set (Fig. S5A). This resulted in the population-average expression of HIPSTR of 0.91 FPKM in these 96 individual K562 cells, which was comparable with ENCODE Project bulk RNA-seq data for K562 cells (Fig. 1G).
The present study and the previously published analyses of transcriptomes of single mouse immune cells10 seem to be in disagreement with a recent work by Cabili et al.11, which shows that lncRNAs and mRNAs have a similar cell-to-cell abundance distribution. We found that 33 out of the 61 lncRNAs reported in ref. 11 were expressed (>3 FPKM) in at least one cell from at least one of the five single-cell RNA-seq data sets analyzed in the present study (Table S10). As anticipated, abundant lncRNAs, such as GAS5 or DANCR, were expressed in the majority of the cells analyzed. On the other hand, in many instances, e.g. lincFOXF1 or lincMKLN1, we found that lncRNAs assayed in Cabili et al.11 were as well expressed at relatively high levels, but only in few cells, and were classified as transcripts with high expression heterogeneity (Table S10). Similar pattern of expression was observed for another lncRNA, linc-MUC20-1 (XLOC_024513, not tested in Cabili et al.11). Heterogeneity of linc-MUC20-1 expression was different in different cell types with comparable expression levels of this lncRNA: low expression heterogeneity in K562 cells (average – 2.88 FPKM, max – 19.91 FPKM, expressed in 27% of cells), and high heterogeneity – in 19 W hPGCs (average – 1.97 FPKM, max – 27.18 FPKM, expressed in 14% of cells). Expression pattern of linc-MUC20-1 therefore demonstrates that expression heterogeneity of a given, readily detectable gene is not a fixed, but rather a very dynamic attribute that depends on a cell type considered for the analysis, which may explain the different conclusions reached by Cabili et al.11.
Based on these data, we conclude that highly heterogeneous expression in a population of seemingly identical cells is a common feature of human lncRNAs. This adds to the previously reported developmental stage, tissue, and cell subtype specificity of lncRNA expression6,7,8,34. It is important that the observed differences in cell-to-cell variation of abundance between lncRNAs and mRNAs cannot be explained by drop-out effects or technical noise of single-cell RNA-seq data, because our comparisons were done only for readily detectable genes (max expression >3 FPKM) with similar abundances within the same data sets.
Our results are of special importance for the studies of numerous recently identified and uncharacterized lncRNAs, as a complete absence of a given lncRNA in multiple cells in a population complicates statistical analyses, and high cell-to-cell variability in lncRNAs levels suggests that analyses of hundreds or even thousands of individual cells might be required to reveal meaningful expression correlations between heterogeneously expressed lncRNAs and other genes. For this, development of reliable and easy-to-use techniques facilitating enrichment for subpopulations of live cells expressing a lncRNA of interest will be required to uncover the exact mechanism of action of heterogeneously expressed lncRNAs, such as HIPSTR.
Methods
Full methods are available on-line in the Supplementary Materials section.
Antisense oligonucleotide (ASO)-mediated silencing
For ASO-mediated silencing of HIPSTR 4.5 × 105 HEK293 cells or 2.4 × 105 LNCaP cells were plated on 6-well plates 24 h before transfection. Transfections were performed by using 0.025 μl of Lipofectamine RNAiMAX (Invitrogen) per 1 pmol of transfected ASO. Transfection mixes were prepared in OptiMEM I Reduced Serum Medium (Gibco). TFAP2A-AS1 silencing in HEK293 cells was done as described above for HIPSTR. To silence HIPSTR expression in H1BP cells, 4 × 104 cells were plated on 6-well plates 48 h before transfection, and cultured as described above; 0.013 μl of GenMute siRNA Transfection Reagent (SignaGen) per 1 pmol of ASO were used for transfection. Transfection mixes were prepared in 1x GenMute Transfection Buffer (SignaGen). A total of 300 pmol of ASO or mix of ASOs per well on 6-well plates was used for transfection. In all silencing experiments cells were collected for subsequent RNA or protein extraction 24 h after transfection with ASOs. For time-course HIPSTR knockdown assay in HEK293 cell line, cells were collected 6, 12, 24, 48, and 72 h after transfection with ASOs.
Oligonucleotide sequences
All oligonucleotide sequences (primers and ASOs) are listed in Table S11.
Microarray experiments
200 ng of total RNA from HEK293 cells or 100 ng of total RNA from H1BP cells transfected with ASOs targeting HIPSTR were converted into Cy3- and Cy5-labeled cRNA with the Agilent Low Input Quick Amp Labeling Two Color Kit. Dye-swap technical replicates were created for each biological replicate. Three biological replicates of HEK293 cells transfected with each ASO were used for microarray experiments. In experiments with H1BP cells, three biological replicates for control ASO, and two – for each of the targeting ASOs were assayed. Obtained cRNA samples were then hybridized to Agilent SurePrint G3 Gene Expression Microarrays (G4851B) 8 × 60 K as per manufacturer’s instructions. Data intensities were extracted from the slide images with Feature Extraction Software (Agilent Technologies) and normalized by using the Lowess method (Agilent Technologies).
All probes whose mean signal was lower than background on at least one array were filtered out. Signal intensities were normalized by 40% trimmed mean. Significance Analysis of Microarrays (SAM) with two-class comparison was then used to identify differentially expressed genes58. SAM q-value ≤ 0.01 and fold change ≥2 were considered as a threshold for identification of differentially expressed genes. Hierarchical clustering of differentially expressed genes was done with TIBCO Spotfire software by applying Z-score transformation of the normalized data intensities for each gene across all samples.
Gene Ontology (GO) analysis
GO and tissue-specific expression analyses of annotated differentially expressed genes were performed with DAVID (https://david.ncifcrf.gov/) (ref. 59) with GOTERM_BP_ALL (or GOTERM_CC_FAT – for genes upregulated upon HIPSTR knockdown in H1BP cells) and UP_TISSUE tables, respectively. Benjamini-Hochberg adjusted p-value ≤ 0.01 was used as a significance threshold. Genes are referred to as “annotated” if they have a HGNC symbol in Agilent annotation.
RNA-seq and ChIP-seq analysis
ENCODE Project29 human long polyadenylated RNA-seq data for the indicated cell lines were obtained from GEO entry GSE30567, and mouse long RNA-seq – from GEO entry GSE36025. Ribosome profiling data from ref. 33 were downloaded from SRA entry SRA099816. K562 single-cell RNA-seq data were downloaded from SRA entry SRX495504 (ref. 53). Human and mouse embryonic single-cell RNA-seq data were retrieved from ENA entry PRJEB8994 (ref. 51), and from GEO entries GSE44183 (ref. 50), GSE36552 (ref. 7), GSE57249 (ref. 52), and GSE63818 (ref. 54). RNA-seq of DRB- (RNA Pol II elongation inhibitor) or vehicle-treated HEK293 cells from ref. 60 were obtained from GEO entry GSE66478. H3K4me3 ChIP-seq data for liver samples of 10 mammalian species were downloaded from Array Express website entry E-MTAB-2633 (ref. 44), for testis samples of mouse and rooster – from GEO entry GSE44588 (ref. 43), for frog blastula, gastrula, neurula and tailbud stage embryos – from GEO entry GSE41161 (ref. 42), and for zebrafish 256 cell, oblong and dome stage embryos – from GEO entry GSE44269 (ref. 45). TFAP2A and H3K4me3 ChIP-seq data for chimpanzee NCCs and hNCCs were obtained from GEO entry GSE70751 (ref. 24).
Sequencing data were preprocessed with Trimmomatic v.0.30 (ref. 61) with parameters -phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15. Trimmomatic parameter MINLEN: was set at 16 for ChIP-seq reads, at 20 – for RNA-seq reads, except for RNA-seq data from ref. 33, for which it was set at 30. Additional clipping of adapter sequence CTGTAGGCACCATCAAT was done for preprocessed RNA-seq reads from ref. 33 with fastx_clipper from FASTX Toolkit v.0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/). Human RNA-seq reads were mapped with TopHat v.2.0.12 (ref. 62) and Bowtie v.2.2.3 (ref. 63). The following parameters for TopHat were used: –no-coverage-search –b2-sensitive; for paired-end strand-specific RNA-seq data (except LNCaP RNA-seq), –library-type fr-firststrand parameter was used in addition to the mentioned above; for LNCaP RNA-seq data –library-type fr-secondstrand parameter was added. ChIP-seq reads were mapped by Bowtie v.2.2.3 with parameter: –sensitive. Read densities were retrieved with genomecov command from bedtools package v.2.20.1 (ref. 64), and UCSC Genome Browser tracks were built with bedGraphToBigWig v.4 (ref. 65). To count RNA-seq reads, TopHat paired-end RNA-seq data alignment output files were first sorted by read names with sort command from SAMtools package v. 0.1.19-44428cd (ref. 66). RNA-seq reads were counted with htseq-count v.0.6.1p1 (ref. 67), with parameter -s yes for single-end strand-specific data sets, -s reverse – for paired-end strand-specific data sets, and -s no for non-stranded data sets. For human data sets we used a GTF file from GSE57049 (ref. 11) complemented with the genomic coordinates of HIPSTR. Gene names in the output tables are those found in the abovementioned GTF file. To count mapped reads in mouse data sets, we used a GTF file for mouse genome assembly mm9 that was fetched from the illumina support site (https://support.illumina.com/). Gene expression levels were calculated in FPKM, considering gene length as a sum of all exonic non-overlapping sequences of all isoforms of a given gene. Unless stated otherwise, ChIP-seq and RNA-seq data are presented as aggregates of biological replicates for each indicated condition to increase resulting genome and transcriptome coverage, respectively.
To map RNA-seq and ChIP-seq data, the following reference genome assemblies were downloaded from UCSC Genome Browser (http://hgdownload.soe.ucsc.edu/downloads.html): galGal4 (chicken), panTro4 (chimpanzee), bosTau7 (cow), canFam3 (dog), xenTro3 (frog), hg19/GRCh37 (human), calJac3 (marmoset), mm9 (mouse), monDom5 (opossum), susScr3 (pig), oryCun2 (rabbit), rn5 (rat), rheMac3 (rhesus), danRer7 (zebrafish).
For single-cell RNA-seq data analyses, genes were considered as protein-coding if they were assigned RefSeq accession prefix NM_ (mRNA), or XM_ (mRNA predicted), or NP_ (peptide), or XP_ (peptide predicted); we also considered a gene as protein-coding in our analyses, if a possibility of read-through from a lncRNA gene to a protein-coding gene existed, according to GTF annotation file. Genes were considered as non-coding if they were assigned RefSeq accession prefix NR_ (ncRNA), or XR_ (ncRNA predicted), or were annotated as lincRNAs in the GTF file from ref. 11. For comparisons of expression profiles of non-coding and protein-coding genes in single cells, we considered only genes generating transcripts with total length of non-overlapping exonic sequences longer than 200 nt.
Expression heterogeneity comparisons with the Finite Mixture model
To evaluate heterogeneity of gene expression in single cells, we used single-cell RNA-seq data sets for totipotent blastomeres from 8-cell and morula-stage human embryos, hESCs (both – from ref. 7), K562 cells (from ref. 53), or hPGCs from 7-week- or 19-week-old male embryos (both – from ref. 54). For each gene in each data set, we calculated the number of cells N, in which a given gene was expressed. The difference in cell-to-cell variability of gene expression was most evident between lncRNAs and mRNAs with max expression 3–30 FPKM in single cells, and thus we considered only genes with expression levels within this range, and did not consider genes whose expression was >30 FPKM in at least one cell of a data set under analysis. We counted a cell as positive for expression of a given gene if the expression level of that gene was >3 FPKM in that cell, which is a 30-fold more stringent cut-off than used in refs 7, 54.
We observed that, when assessed for all genes, the distribution of their corresponding N values is a mixture distribution. We used the normalmixEM function from mixtools v.1.0.4 R package68 to fit a model mixture distribution with two populations of genes – those with high or low heterogeneity of expression. Parameters used were: number_of_components = 2, lambda = 0.5, sigma = 0.5.
We next applied the resultant model to calculate the posterior probability of each gene under analysis to belong to either the high or the low heterogeneity of expression population. If a given gene could be associated with one of the abovementioned populations with a posterior probability >0.99, it was assigned the “H” or “L” flag (for high or low heterogeneity of expression, respectively; Tables S4,S5,S6,S7,S8), otherwise the “U” (uncertain heterogeneity) flag was assigned.
Additional Information
Accession codes: The microarray data sets supporting the results of this article were deposited in NCBI, and are available in the Gene Expression Omnibus (GEO) repository, GSE77937. RNA-seq data from LNCaP prostate cancer cell line were deposited in NCBI, and are available in the GEO repository, GSE79301. HIPSTR sequence is deposited in GenBank with accession number KU904338.
How to cite this article: Yunusov, D. et al. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci. Rep. 6, 32753; doi: 10.1038/srep32753 (2016).
References
Clark, M. B. et al. The reality of pervasive transcription. PLoS Biol 9, e1000625; discussion e1001102 (2011).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Berretta, J. & Morillon, A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 10, 973–982 (2009).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47, 199–208 (2015).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17, 47–62 (2015).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 (2011).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20, 1131–1139 (2013).
Liu, S. J. et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome biology 17, 67 (2016).
Dinger, M. E., Amaral, P. P., Mercer, T. R. & Mattick, J. S. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic 8, 407–423 (2009).
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome biology 16, 20 (2015).
Nakaya, H. I. et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome biology 8, R43 (2007).
Tahira, A. C. et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 10, 141 (2011).
Fachel, A. A. et al. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: emerging functional associations. Mol Cancer 12, 140 (2013).
Ayupe, A. C. et al. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic regions of the human genome. RNA Biol 12, 877–892 (2015).
Louro, R. et al. Androgen responsive intronic non-coding RNAs. BMC Biol 5, 4 (2007).
Engelhardt, J. & Stadler, P. F. Evolution of the unspliced transcriptome. BMC evolutionary biology 15, 166 (2015).
Beckedorff, F. C. et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet 9, e1003705 (2013).
Magistri, M., Faghihi, M. A., St Laurent, G. 3rd & Wahlestedt, C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet 28, 389–396 (2012).
Pellikainen, J. M. & Kosma, V. M. Activator protein-2 in carcinogenesis with a special reference to breast cancer–a mini review. Int J Cancer 120, 2061–2067 (2007).
Zhang, J. et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381, 238–241 (1996).
Schorle, H., Meier, P., Buchert, M., Jaenisch, R. & Mitchell, P. J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235–238 (1996).
Rada-Iglesias, A. et al. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648 (2012).
Prescott, S. L. et al. Enhancer Divergence and cis-Regulatory Evolution in the Human and Chimp Neural Crest. Cell 163, 68–83 (2015).
Aghajanova, L. et al. Comparative transcriptome analysis of human trophectoderm and embryonic stem cell-derived trophoblasts reveal key participants in early implantation. Biol Reprod 86, 1–21 (2012).
Cheng, Y. H. et al. Critical role for transcription factor AP-2alpha in human trophoblast differentiation. Physiol Genomics 18, 99–107 (2004).
Makhov, P. B. et al. Reversal of epigenetic silencing of AP-2alpha results in increased zinc uptake in DU-145 and LNCaP prostate cancer cells. Carcinogenesis 32, 1773–1781 (2011).
Ruiz, M. et al. Activator protein 2alpha inhibits tumorigenicity and represses vascular endothelial growth factor transcription in prostate cancer cells. Cancer Res 64, 631–638 (2004).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15, 7–21 (2014).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35, W345–349 (2007).
Wang, L. et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41, e74 (2013).
Stumpf, C. R., Moreno, M. V., Olshen, A. B., Taylor, B. S. & Ruggero, D. The translational landscape of the mammalian cell cycle. Mol Cell 52, 574–582 (2013).
Ravasi, T. et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16, 11–19 (2006).
Rands, C. M., Meader, S., Ponting, C. P. & Lunter, G. 8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet 10, e1004525 (2014).
Kapusta, A. & Feschotte, C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet 30, 439–452 (2014).
Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).
Necsulea, A. et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014).
Marques, A. C. et al. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome biology 14, R131 (2013).
Schneider, R. et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6, 73–77 (2004).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
van Heeringen, S. J. et al. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res 24, 401–410 (2014).
Li, X. Z. et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50, 67–81 (2013).
Villar, D. et al. Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015).
Zhang, Y. et al. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24, 260–266 (2014).
Guo, G. et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18, 675–685 (2010).
Luscher, B., Mitchell, P. J., Williams, T. & Tjian, R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev 3, 1507–1517 (1989).
Berlato, C. et al. Alternative TFAP2A isoforms have distinct activities in breast cancer. Breast Cancer Res 13, R23 (2011).
Yang, Y. et al. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proceedings of the National Academy of Sciences of the United States of America 112, E2337–2346 (2015).
Xue, Z. et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013).
Tohonen, V. et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun 6, 8207 (2015).
Biase, F. H., Cao, X. & Zhong, S. Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res 24, 1787–1796 (2014).
Luo, H., Li, J., Chia, B. K., Robson, P. & Nagarajan, N. The importance of study design for detecting differentially abundant features in high-throughput experiments. Genome biology 15, 527 (2014).
Guo, F. et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 161, 1437–1452 (2015).
Lin, N. et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53, 1005–1019 (2014).
Pelechano, V. & Steinmetz, L. M. Gene regulation by antisense transcription. Nat Rev Genet 14, 880–893 (2013).
Tsai, M. C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America 98, 5116–5121 (2001).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009).
Werner, M. S. & Ruthenburg, A. J. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep 12, 1089–1098 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S. & Karolchik, D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Benaglia, T., Chauveau, D., Hunter, D. R. & Young, D. S. mixtools: An R package for analyzing finite mixture models. J Stat Softw 32, 1–29 (2009).
DeOcesano-Pereira, C. et al. Long non-coding RNA INXS is a critical mediator of BCL-XS induced apoptosis. Nucleic Acids Res 42, 8343–8355 (2014).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Acknowledgements
The authors are grateful to Eduardo M. Reis and Ana C. Ayupe for sharing RNA samples extracted from HeLa cells, for the HIPSTR biogenesis assays. We thank Felipe C. Beckedorff and Murilo S. Amaral for the generation of LNCaP RNA-seq data. We also thank Ying Yang and Megan Sheridan for the help with H1BP cells, Sara L. Prescott and Rajini Srinivasan for the help with neural crest cells derivation, and Bianca Dazzani for the help with routine experiments in the lab. This work was funded by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant 2014/03620-2) [to SVA], and by NIH grants U01 DE024430 [to JW], HD-067759 [to RMR], and HD-077108 [to TE]; DY and LA received fellowships from FAPESP, and LFDS received a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). SVA received an established investigator fellowship award from CNPq.
Author information
Authors and Affiliations
Contributions
D.Y. and S.V.-A. conceived the project, designed experiments, analyzed the data, and wrote the manuscript, with input from all authors. D.Y. carried out all experiments, and RNA-seq and ChIP-seq data mining and analyses; L.A. provided help with microarray and western blot experiments and analyses; L.F.D.S. performed random shuffling of HIPSTR sequence, and performed the analyses of expression heterogeneity. J.W. contributed reagents and provided expertise in experiments with hNCCs. T.E. and R.M.R. contributed reagents and provided remarkable expertise in experiments with H1BP cells.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yunusov, D., Anderson, L., DaSilva, L. et al. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci Rep 6, 32753 (2016). https://doi.org/10.1038/srep32753
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32753
This article is cited by
-
FGD5-AS1 promotes growth and mobility of trophoblast cells by regulating IGF2 via sponging miR-16-5p in pre-eclampsia
Molecular & Cellular Toxicology (2023)
-
Transcriptome Sequencing Identifies Potential Biomarker for White Matter Lesions Diagnosis in the Hypertension Population
Neurochemical Research (2021)
-
The lncRNA Fer1L4 is an adverse prognostic parameter in clear-cell renal-cell carcinoma
Clinical and Translational Oncology (2020)
-
BcMF11 and its homologous sequences may form a lncRNA family in Brassica diploids
Acta Physiologiae Plantarum (2018)
-
Identification of long non-coding RNA in the horse transcriptome
BMC Genomics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.