Abstract
Unreduced egg formation (apomeiosis) in flowering plants is rare except when it is coupled with parthenogenesis to yield gametophytic apomixis via apospory or diplospory. Results from genetic mapping studies in diverse apomictic taxa suggest that apomeiosis and parthenogenesis are genetically linked, a finding that is compatible with the conventional rationale that apomeiosis is unlikely to evolve independently because of deleterious fitness consequences. An Erigeron annuus (apomictic) × E. strigosus (sexual) genetic mapping population, however, included a high proportion of plants that were highly apomeiotic (diplosporous) but nonapomictic; that is, they lacked autonomous seed production. To evaluate the function and inheritance of diplospory in Erigeron, a diplosporous triploid (2n=3x=27) seed parent was crossed with a sexual diploid (2n=2x=18) E. strigosus pollen parent to produce an F1 of 31 plants. Chromosome numbers and molecular markers (AFLPs) document the inheritance of the maternal genome through unreduced eggs resulting in recombinant but predominantly (77%) tetraploid F1s (2n=4x=36; 2n+n, BIII). Quantitative evaluation shows continuous variation in the proportion of diplosporous (vs meiotic) ovules (41–89%) in tetraploid F1s despite the presumed equal genetic contribution from the diplosporous mother. These findings demonstrate the functional independence of diplospory and suggest that variation in the trait in F1s is likely due to segregating paternal modifiers. In addition, of six aneuploid (4x−1, 4x−2) F1s, three lack a subset of maternal AFLP markers. These plants likely arose from aberrant megagametogenesis resulting in the loss of maternal chromatin prior to fertilization.
Similar content being viewed by others
Introduction
In naturally occurring sexual plants, the formation of unreduced megagametophytes, or apomeiosis, is infrequent except in ephemeral interspecific hybrids. None-the-less, apomeiosis is considered to be a key step in polyploid evolution (reviewed in Ramsey and Schemske, 1998). In contrast, apomeiosis achieves high and regular expression in species that reproduce via gametophytic apomixis (Nogler, 1984a; Asker and Jerling, 1992). In apomicts, the unreduced megagametophyte is formed apomeiotically via either apospory (the gametophyte arises from a cell of the ovule other than the megaspore mother cell (MMC)) or diplospory (the gametophyte arises directly from the MMC). However, maternal ploidal level is maintained across generations because the unreduced egg borne by the megagametophyte develops parthenogenetically into an embryo, and progeny are thus genetically identical to the mother plant (barring chiasmata prior to restitution). In apomicts, the nutritive endosperm may also form autonomously, or fertilization of the polar nuclei by a sperm cell may be required (pseudogamy).
Genetic mapping studies in diverse taxa suggest that apomeiosis and parthenogenetic embryo formation are genetically linked (see reviews by Savidan, 2000; Grossniklaus et al, 2001). According to Mogie (1992), this result is explained by a single gene model for apomixis in which parthenogenesis occurs as a pleiotropic consequence of apomeiosis. This view is furthered by logical arguments that apomeiosis and parthenogenesis are unlikely to evolve independently because each trait, separately, would be deleterious (Asker and Jerling, 1992). This model is controversial, however, because apomixis often maps to regions with low recombination. Therefore, it is possible that these apomixis regions harbor many major or modifying genes affecting distinct aspects of the trait.
Curiously, fertilization of unreduced ovules has often been recorded as a source of 2n+n (or BIII) ‘off types’ in progeny tests of apomictic plants (Asker and Jerling, 1992). This phenomenon demonstrates that unreduced eggs produced by apomicts can function normally in crosses. However, our knowledge of the function and inheritance of apomeiotic ovules is limited because few apomeiotic-only plants have been observed in nature. For instance, only two known examples of naturally occurring apomeiotic plants (in Parthenium and Potentilla) are cited in Asker and Jerling (1992; p 82).
In contrast, apomeiotic plants occur in relatively high frequency in some recently reported crossing experiments. In these studies, F1 populations resulting from sexual × apomict crosses include plants that produce unreduced ovules in the absence of other components of apomixis. In aposporous Poa pratensis (Poaceae), for example, apomeiotic individuals (identified cytologically) constituted 5% of F1s (Albertini et al, 2001). In diplosporous Taraxacum officinale (Asteraceae), six of 19 F1s (32%) were identified as apomeiotic, producing at least some 2n+n and no 2n+0 offspring progeny when test-crossed (van Dijk et al, 1999). In Erigeron (Asteraceae), diplosporous plants constituted 23% (30/130) of an F1 mapping population generated by crossing apomictic E. annuus with sexual E. strigosus. In most of the diplosporous plants, both reduced and unreduced ovules coexisted in the same plant (the proportion of unreduced ovules ranging from 8 to 100%), suggesting that penetrance of apomeiosis may be influenced by modifiers or genetic balance (Noyes, 2000). Together, these three studies support the hypothesis that apomeiosis may be distinct from other aspects of apomictic development (see review by Grossniklaus et al, 2001). This view is supported by genetic mapping in Erigeron that shows that diplospory and autonomous seed formation map to separate linkage groups (Noyes and Rieseberg, 2000), and developmental analysis for Tripsacum dactyloides that indicate no functional relationship between diplospory and parthenogenesis (Grimanelli et al, 2003).
In this work, the function of apomeiotic (diplosporous) ovules and the inheritance of the trait in Erigeron are considered. A highly diplosporous seed parent was crossed with a sexual diploid to test the function of unreduced ovules. The contributions of paternal and maternal genomes in F1s were evaluated using molecular markers, and the inheritance of diplosporous ovule production was assessed by comparing levels of diplospory, measured quantitatively, in the maternal plant and F1s.
Materials and methods
A triploid (2n=27; Figure 1) diplosporous plant lacking seed production (H131) was selected that was a member of a mapping population resulting from a cross between triploid apomictic E. annuus and sexual diploid E. strigosus (Noyes, 2000). This plant was selected because it produced 92% unreduced ovules (Figure 2). Of its remaining ovules, 3% were apparently meiotic, yielding four approximately equal sized, but likely chromosomally unbalanced, nuclei from divisions of the MMC. The remaining 5% of ovules were aberrant, exhibiting during early megagametogenesis either (1) two relatively large nuclei, as in diplosporous ovules, plus one or more very small nuclei (micronuclei; Figure 2), or (2) three approximately equal sized nuclei of uncertain origin but likely resulting from a combination of restitution and reduction following meiosis I. In both aberrant types, nonmaternal (but abnormally reduced) eggs would likely result. Mature unreduced megagametophytes of H131 include a normal egg apparatus consisting of two synergids and egg cell, and a central cell containing a single large nucleus inferred to be derived from the fusion of two polar nuclei (the fusion nucleus; Figure 2). H131 exhibits no sign of autonomous embryo or endosperm formation. This plant served as seed parent in a cross with a field collected wild-type sexual diploid E. strigosus (2n=18; Figure 1) pollen donor (Noyes #1316) from Marion, Co., Georgia, USA. This plant exhibited high quality pollen (90% estimated viability when stained in Cotton Blue in Lactophenol), ovule development was determined to be exclusively sexual by means of tetraspory (Figure 2), and mature megagametophytes were similar in structure to those produced by the apomeiotic seed parent. Eight capitula of H131were pollinated over a 5-day period and mature heads were collected after approximately 2 weeks. The cross yielded more than 1200 achenes and after 3 weeks storage at 4°C, 50 achenes were germinated in a Petri dish. A total of 31 individuals that survived seedling stage were grown to maturity in a University of Colorado greenhouse.
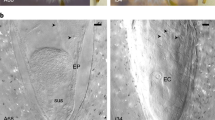
Micrographs of cleared Erigeron megagametophytes. (a) Young sexual (tetrasporic) gametophyte showing four nuclei resulting from meiosis of MMC. (b) Young apomeiotic (diplosporous) gametophyte showing two nuclei resulting from mitotic (or mitotic-like) division of MMC. (c) Aberrant gametophyte showing two large nuclei (as in apomeiotic gametophytes) plus two micronuclei. (d) Mature reduced megagametophyte. (e) Mature unreduced megagametophyte. Micropyle is at the base of each micrograph. en=egg nucleus, mn=micronucleus, n=nucleus, nc=nucellus, fn=fusion nucleus. Bar=10 μm.
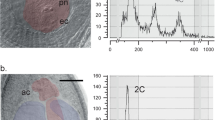
Root tips were collected from each F1 prior to bolting, and chromosome counts were made using acetocarmine squash methods and light microscopy detailed previously (Noyes, 2000). In total, five to eight well-separated spreads were examined at × 1000 using an Olympus BX51 light microscope to ensure accurate chromosome number determination.
All F1s flowered after approximately 6 months. To evaluate ovule development, young capitula in bud stage were fixed in FAA, stored in 70% ethanol, and subsequently cleared in methyl salicylate following serial dehydration in ethanol as previously described (Noyes, 2000). Ovule development in parentals and the 31 F1s was evaluated quantitatively for 50–65 each of ray and disc florets per individual, using an Olympus BX51 microscope equipped with differential interference contrast (DIC) optics. Both types of florets were examined in the case that ovule development might be different in the two floret types or, alternatively, in peripheral vs centrally located florets. Ovules were scored as diplosporous (two nuclei resulting from the division of the MMC), meiotic (four nuclei resulting from the division of the MMC), or as aberrant (see above). Final scores for individual capitula were calculated with equal weighting of separate ray and disc measures. To evaluate variation among capitula of an individual, ovules from two additional heads were evaluated for nine F1s. Variation between ray and disc florets within a single capitulum and among capitula was evaluated statistically using nonparametric and ANOVA analyses. Ovule and chromosome images were captured and manipulated using AnalySIS (v. 3.1) image processing software (Soft Imaging System, GmbH 1989–2001) and Adobe Photoshop (version 6.0.1, Adobe Systems, Inc., San Jose, CA, USA).
DNA was isolated from parents and F1s using the DNeasy Plant Mini DNA extraction kit (Qiagen, Valencia, CA, USA) and quantified using a Hoefer DyNA Quant 200 Fluorometer (Amersham Pharmacia Biotech, San Francisco, CA, USA). To evaluate inheritance patterns, Amplified Fragment Length Polymorphisms (AFLPs) (Vos et al, 1995) were evaluated. Polymorphisms from six selective primer combinations (Eacc × Maac, Eacc × Macc, Eacc × Macg, Eacc × Magc, Eacg × Macg, Eacg × Magc) were generated using standard methods (Noyes and Rieseberg, 2000), and PCR products were resolved on an MJResearch (Waltham, MA, USA) BaseStation that utilizes a 6% ultrathin acrylamide gel and hardware permitting detection of EcoRI selective primers 5′ labeled with FAM, TET, and HEX. ROX-labeled standard size marker (MegaBace ET-900, Amersham Pharmacia Biotech, San Fransisco, CA, USA) was included in each sample to facilitate interlane comparisons. Gels were run for approximately 5 h at 17 000 V, which permitted evaluation of bands to 500 bp. Gels were evaluated using Cartographer (v. 1.06, MJResearch, Waltham, MA, USA). Informative AFLP loci were identified as those unique to either parent, and the software was used to demarcate peak ranges for each locus. Each locus was treated as dominant. Individuals were scored manually by scrolling through multielectropherogram displays. Loci were evaluated for mode of inheritance and segregating patterns were subjected to χ2 test of Mendelian inheritance. Gel images were exported as TIFF files and manipulated in Adobe Photoshop.
Results
Chromosome numbers of F1s (Figure 1) were predominantly tetraploid (2n=36) (24/31; 77%), but six of 31 F1s (19%) were 2n=35 or 2n=34, and one F1 was 2n=23. The cytotypes of parentals were observed to consist of metacentric and submetacentric chromosomes of approximately equal length and no extraordinarily large or small chromosomes were observed among the F1s.
For F1s, disc florets consistently exhibited a greater percentage of diplosporous ovules than ray florets within the same capitulum (30/31 F1s, sign test P=1.44 × 10−8), with an individual on average exhibiting 18% greater proportion of diplosporous ovules in the disc compared to the rays (range=−0.4 to 72.0%). Concomitantly, capitula exhibited lower proportions of both meiotic and aberrant ovules in the disc compared to the rays (27/31 and 28/31 F1s, sign test P=1.46 × 10−5 and 2.09 × 10−6, respectively).
Diplospory estimates varied considerably among F1s, even among F1s of a single chromosome number (Figure 3). Among tetraploid (2n=36) F1s, mean diplospory values ranged from 41 to 89%. In addition, replicate measures of three capitula for four individuals representing a range of values of diplospory in tetraploids showed that there is only relatively modest within-individual variation (Figure 3). ANOVA among these four tetraploids supports the hypothesis that most of the variation is among rather than within individuals (P =5.4 × E × 1010). Multiple estimates of diplospory for selected individuals with 2n=35, 2n=34 also showed high variance among individuals within ploidal level, but low levels of within-plant variation (Figure 3). None-the-less, while there is an experiment-wide average decrease in mean diplospory for all F1s compared to their mother (67.1 vs 92.1%), there was no apparent relationship between F1 diplospory level and decreasing chromosome number.
The six AFLP primer combinations yielded a total of 82 maternal and 39 paternal informative loci (Table 1; Figure 4). Of the 82 maternal bands, 81 are present in all 24 2n=36 progeny, as would be expected for strict inheritance of the entire maternal genome; one tetraploid F1 lacked one band. In contrast, three and eight maternal markers were missing in two of the four 2n=35 progeny, one marker was absent in one 2n=34 F1, and 29 markers were absent in the 2n=23 F1. The absence of evidence of marker loss in three of the seven aneuploids was unexpected and was initially attributed to erroneous chromosome number determination. However, sufficient material permitted reanalysis of one of the presumed aneuploids, for which 2n=35 was reconfirmed. All markers were anonymous with respect to phenotype except for three maternal AFLP markers known to be linked to diplospory (Eacg × Magc.340, Eacg × Magc.350, Eacc × Macg.170; Noyes and Rieseberg, 2000). All progeny, regardless of chromosome number, inherited these bands. In total, 50 of 82 maternal bands (61%) were invariant among all progeny.
Inheritance of AFLP markers in apomeiotic Erigeron. Maternal ‘M’, paternal ‘P’, and F1 bands (individuals 1–31) as indicated. F1s 1–24 are tetraploid (2n=36), F1s 25–28 are aneuploid (2n=35), F1s 29–30 are aneuploid (2n=34), F1 31 is 2n=23. (a) Maternal band M.Eacg × Magc.340 linked to diplospory is present in all F1s. (b) Maternal band M.Eacg × Magc.393 segregates among aneuploids. Arrows indicate two individuals lacking bands. (c) Paternal band P.Eacc × Macg.085 is present in all F1s (band for individual 18 is faint). (d) Paternal band P.Eacg × Macg.250 segregates among F1s. Images from ‘unsmiled’ TIFF gel images exported from Cartographer (MJResearch, Inc.).
In contrast, only six of 39 paternal bands (15%) were monomorphic for F1s (Table 1; Figure 4). These loci demonstrate that each F1 received a genetic contribution from the pollen parent and that all F1s are therefore recombinant. The remaining 33 loci segregated among the 31 progeny. Although the mean observed number of markers across F1s (16.1) was slightly greater than the expected value of 15.5, segregation patterns did not differ significantly from Mendelian expectations (P>0.05, α=0.0016, Bonferroni corrected for multiple tests).
Discussion
Results unequivocally demonstrate that unreduced (2n) and aberrant (2n−1, 2n−2) egg cells produced by diplosporous (but nonapomictic) Erigeron function in crosses. The majority of F1s (77%) show an increase in chromosome number from 2n=27 to 2n=36 that is consistent with 2n+n (BIII) hybridization. In addition, AFLP markers show that (1) tetraploid F1s inherited the complete maternal nuclear genome (except for one individual for one locus), and (2) the diploid pollen parent contributed genetically to each F1. This observation is consistent with results obtained from test-crosses of apomeiotic segregants in Taraxacum (van Dijk et al, 1999). These experiments demonstrate that apomeiotic individuals can have full reproductive function through unreduced gametes. In addition, the F1 with 2n=23, assumed to combine 14 maternal and nine paternal chromosomes via n+n (=BII) hybridization, indicates that reduced egg cells in apomeiotic Erigeron may also function in crosses.
These results support the hypothesis that apomeiosis may be functionally and genetically independent from other aspects of apomixis (parthenogenesis, autonomous endosperm formation), but begs the question of how the trait, which by itself should lead to genetic collapse through recurrent polyploidization, could possibly evolve. One possible solution is in the observation that in Erigeron, autonomous seed formation appears to be contingent upon diplospory; that is, genes linked to autonomous seed formation appear to be silent (at diploid and polyploid levels) in the absence of apomeiosis (Noyes and Rieseberg, 2000). A similar conclusion has been inferred for aposporous P. pratensis (Albertini et al, 2001). This suggests an evolutionary model in which genes capable of causing autonomous seed formation, possibly neutral in the absence of apomeiosis, may reside silently in the genomes of sexual plants. Subsequent fortuitous mutations or gene interactions in hybrid plants, which result in apomeiosis, could then function cooperatively with the previously inactive seed genes to yield apomixis. Accordingly, the separate elements of apomixis (apomeiosis, parthenogenesis, and autonomous or pseudogamous endosperm formation) need not evolve independently, thus overcoming a principal criticism of multigene models for apomixis (Asker and Jerling, 1992, pp. 83). The mechanism by which a gene for autonomous seed formation might function and interact with a locus for apomeiosis is completely unknown at this time.
Aneuploids with 2n=35 (4x−1) and 2n=34 (4x−2), constituting 19% of F1s, have evidently arisen via loss of maternal chromosomes prior to fertilization of the egg cell. This result is consistent with the observation of aberrant megagametophytes (5%) in the apomeiotic seed parent. Loss of maternal chromatin is further evidenced by the absence of maternal AFLP markers in three of the six aneuploid F1s. Marker absence, plus no evidence of extraordinarily large chromosomes, demonstrates that the reduction in number of chromosomes of maternal origin is not due to chromosomal fusion.
In analyses of apomictic sib-families in diverse taxa, individual progeny are typically classified as either (1) maternal through strict apomixis (2n+0), (2) polyhaploid, that is, arising parthenogenetically from reduced eggs (n+0), (3) recombinant via unreduced eggs (2n+n, BIII), (4) recombinant via reduced eggs (n+n, BII), or, more rarely (5) maternal variant arising through automixis (Nybom, 1996; Savidan, 2000). The origin of aneuploids through aberrant megagametogenesis in apomicts is rarely recorded, although has been well documented in diplosporous Elymus rectisetus (Hair, 1956; Crane and Carman, 1987) and Taraxacum officinale (Richards, 1996). Chromosome loss in these taxa is attributed to incomplete restitution, partial segregation, and chromosomal nondisjunction in early megagametogenesis. According to Asker and Jerling (1992, p 121), this mode constitutes a form of autosegregation. Aneuploids among progeny of apomeiotic Erigeron likely arise by a mechanism similar to that observed in Elymus and Taraxacum. Although not reported for Erigeron in nature, cytological analysis and progeny tests show that formation of micronuclei during megagametogenesis and the subsequent formation of occasional aneuploid progeny is a common feature of apomictic hybrids under experimental conditions (Noyes, 2000; Noyes, unpublished). No doubt this phenomenon will also eventually be found in natural populations.
The differences between ray and disc flowers in expression of diplospory suggest the presence of a gradient influencing ovule development across the Erigeron capitulum. However, because disc flowers were sampled without regard to position, it is not possible to determine if results reflect discontinuous differences between ray and disc flowers, or a more continuous pattern from central to peripheral capitulum regions. Differential homeotic gene expression based on positional effects in the Asteraceae capitulum has been hypothesized to account for discrete differences in ray vs disc flower morphology (Yu et al, 1999; Gillies et al, 2002; Dezar et al, 2003) and it is plausible that a gene directing the shift from meiotic to diplosporous ovule development is also expressed spatially or along a gradient.
There is a high level of variation among tetraploid F1s in diplospory (Figure 3) despite the fact that each has presumably inherited the exact same capacity for diplospory, including markers linked to the trait, from their triploid mother. Variation in expression of apospory and diplospory within a single genotype has been investigated in several apomictic grass taxa (see Nogler, 1984a), and is shown to be influenced principally by day length and temperature. Environment is likely not the origin of variation in this experiment because capitula from all individuals were all collected within a 5-week period in spring 2002 in a greenhouse under uniform light and temperature regimes.
The most likely explanation is that variation among F1 BIIIs is due to the segregating contribution of the heterozygous pollen parent. The resulting genetic interactions with the presumably constant maternal genome are thus probably responsible for the great range of variation in diplospory (42–89%) among the F1s. This result suggests that the expression of diplospory is not genetically robust (sensu de Visser et al, 2003). Instead, the trait appears to be strongly affected by genetic environment. While it is conceivable that the variation is due to differential epigenetic or maternal effects upon the genetically uniform cohort of egg cells, this is considered to be unlikely. Nonetheless, it would be informative to quantitatively evaluate the inheritance of diplospory in individual apomictic plants that produce both reduced and unreduced ovules. In this way, maternal effects on diplospory would be isolated because there would be no paternal genetic contribution across generations. Also, experiments in which multiple diplosporous plants are crossed with different pollen donors could provide insight into variation in paternal genetic effects.
These results for diplosporous Erigeron are anticipated in part by work in aposporous Ranunculus auricomus in which variation in BIII progeny were observed across generations of increasing ploidal level (Nogler, 1984b). Although sample sizes of BIIIs were relatively small (seven individuals each at the tetraploid and pentaploid levels), and the BIIIs were pooled from mothers with different genotypes and reproductive phenotypes, R. auricomus none-the-less demonstrates both a quantitative drop in apomeiosis with the addition of sexual genotypes as well as variation in apospory among BIIIs in each generation (triploid F1s: 25–55%, tetraploid BIIIs: 10–20%, pentaploid BIIIs: 5–10%). The continuous variation observed in Erigeron furthermore is consistent with the presence of multiple segregating modifiers of small effect. Lastly, differing genetic interactions in F1s are likely responsible for variation in proportion of ovules (1–15%) exhibiting aberrant division of the MMC.
The overall variation among F1s in ovule development could be due to modifier loci acting directly on diplospory gene expression, changes in modifier dosage, novel epistatic interactions, or a combination of effects. Modifiers have been invoked to explain different developmental patterns (timing of initiation, position, developmental progression) of aposporous embryo sacs of experimental Hieracium apomicts (Koltunow et al, 2000). Since quantitative assessment of apomeiosis is rarely performed, the capacity of the trait to vary among individuals and the likely influence of genetic modifiers on the trait may be underappreciated. As we accelerate towards elucidating the principal pathways and genes involved in apomixis, the role of modifiers will likely take on greater importance.
References
Albertini E, Porceddu A, Ferranti F, Reale L, Baraccia G, Romano B et al (2001). Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sexual Plant Reprod 14: 213–217.
Asker SE, Jerling L (1992). Apomixis in Plants. CRC Press: Boca Raton, Florida, USA.
Crane CF, Carman JG (1987). Mechanisms of apomixis in Elymus rectisetus from eastern Australia and New Zealand. Am J Bot 74: 477–496.
de Visser AJGM, Hermission J, Wagner GP, Meyers LA, Bagheri-Chaichian H, Blanchard JL et al (2003). Perspective: evolution and detection of genetic robustness. Evolution 57: 1959–1972.
Dezar CA, Tioni MF, Gonzalez DH, Chan RL (2003). Identification of three MADS-box genes expressed in sunflower capitulum. J Exp Bot 54: 1637–1639.
Gillies ACM, Cubas P, Coen ES, Abbott RJ (2002). Making rays in the Asteraceae: genetics and evolution of radiate versus discoid flower heads. In: Cronk QCB, Bateman RM, Hawkins JA (eds) Developmental Genetics and Plant Evolution. Taylor and Francis: New York, pp 233–246.
Grimanelli D, Garcia M, Kaszas E, Perotti E, Leblanc O (2003). Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165: 1521–1531.
Grossniklaus U, Nogler GA, van Dijk PJ (2001). How to avoid sex: the genetic control of gametophytic apomixis. Plant Cell 13: 1491–1497.
Hair JB (1956). Subsexual reproduction in Agropyron. Heredity 10: 129–160.
Koltunow AM, Johnson SD, Bicknell RA (2000). Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sexual Plant Reprod 12: 253–266.
Mogie M (1992). The Evolution of Asexual Reproduction in Plants. Chapman and Hall: London.
Nogler GA (1984a). Gametophytic apomixis. In: Johri BM (ed) Embryology of Angiosperms. Springer-Verlag: Berlin, pp 475–518.
Nogler GA (1984b). Genetics of apospory in apomictic Ranunculus auricomus. V. Conclusion. Bot Helv 94: 411–422.
Noyes RD (2000). Segregation for diplospory and seed production in sexual × agamospermous Erigeron hybrids. Int J Plant Sci 161: 1–12.
Noyes RD, Rieseberg LH (2000). Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155: 379–390.
Nybom H (1996). DNA fingerprinting – a useful tool in the taxonomy of apomictic plant groups. Folia Geobot Phytotax 31: 295–304.
Ramsey J, Schemske DW (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Systemat 29: 467–501.
Richards AJ (1996). Genetic variability in obligate apomicts of the genus Taraxacum. Folia Geobot. Phytotax 31: 405–414.
Savidan Y (2000). Apomixis: Genetics and breeding. Plant Breeding Rev 18: 13–86.
van Dijk PJ, Tas ICQ, Falque M, Bakx-Schotman T (1999). Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83: 715–721.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M et al (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414.
Yu DY, Kotilainen M, Pollanen E, Mehto M, Elomaa P, Helariutta Y et al (1999). Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J 17: 51–62.
Acknowledgements
The author thanks three anonymous reviewers for helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Noyes, R. Inheritance of apomeiosis (diplospory) in fleabanes (Erigeron, Asteraceae). Heredity 94, 193–198 (2005). https://doi.org/10.1038/sj.hdy.6800597
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800597
Keywords
This article is cited by
-
Small RNA sequencing provides candidate miRNA-target pairs for revealing the mechanism of apomixis in Zanthoxylum bungeanum
BMC Plant Biology (2021)
-
Ploidy dependent expression of apomixis and its components in guinea grass (Panicum maximum Jacq.)
Euphytica (2018)
-
Morphological, cytological and reproductive characterization of tri-species hybrids (GOS) between Pennisetum glaucum, P. orientale and P. squamulatum
Euphytica (2010)
-
Apomeiotic pollen mother cell development in the apomictic Boechera species
Biologia plantarum (2009)
-
Gamete formation without meiosis in Arabidopsis
Nature (2008)