Abstract
Mutations in the NHS gene cause Nance–Horan Syndrome (NHS), a rare X-chromosomal recessive disorder with variable features, including congenital cataract, microphthalmia, a peculiar form of the ear and dental anomalies. We investigated the NHS gene in four additional families with NHS from the Netherlands, by dHPLC and direct sequencing. We identified an unique mutation in each family. Three out of these four mutations were not reported before. We report here the first splice site sequence alteration mutation and three protein truncating mutations. Our results suggest that X-linked cataract and NHS are allelic disorders.
Similar content being viewed by others
Introduction
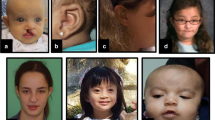
Nance–Horan syndrome (NHS) (MIM♯ 302350), also referred to as cataract–dental syndrome, is a rare X-linked disorder. In hemizygous males, NHS is characterised by severe congenital dense nuclear cataracts, microcornea, multiple dental anomalies, characteristic facial features and in about 30% of the cases developmental delay.1, 2, 3, 4, 5 Carrier females show only mild and variable symptoms of the disease.
Using linkage analysis, the gene for NHS was previously localized to Xp22.31–p22.13 by several groups.6, 7, 8, 9 More recently, Burdon et al10 confirmed this position and identified the gene for NHS by positional cloning. The NHS gene codes for a 1630-amino-acid putative nuclear protein, which might have a function in the regulation of tooth, eye, brain and craniofacial development.10, 11 So far, up to eight protein truncation mutations have been identified in several domains of the NHS gene.10, 11, 12
X-linked cataract (XLC) is one of the clinical features of NHS. Both the NHS and XLC disease genes are localized to the same X-chromosomal region. It is currently not clear whether or not isolated XLC and NHS are allelic disorders. Brooks et al11 studied a family with isolated X-linked congenital nuclear cataract (CXN), but did not identify mutations in the NHS gene. In addition, Burdon et al10 analysed a family with typical NHS symptoms but was also not able to identify the mutation. These cases illustrate that yet undiscovered mutations in intronic or regulatory NHS gene sequences may exist. Alternatively, NHS and XLC may be genetically heterogeneous.
In this study, we (re-)investigate four families with NHS from the Netherlands. These include families previously studied by van Dorp and Delleman4 and Bergen et al.7
We report here the first splice site sequence alteration mutation in the NHS gene and three protein truncating mutations. Three of these pathogenic variants are novel, while the fourth known mutation was previously described as a de novo mutation identified in a single family member.10
Methods
Approval for this study was obtained from the Ethical committee of the Academic Medical Centre, Amsterdam, the Netherlands. Written informed consent was obtained from all participating individuals or their guardians.
All four NHS families were collected from the ‘Netherlands National Archive of Genetic Eye diseases’ and displayed typical features of the syndrome (see Table 1). All available family members were examined by one or more ophthalmologists (NT and or LP). Standard physical examination and standard funduscopy including fundus and lens pictures were carried out. No special equipment or techniques were necessary to provide a sound diagnosis.
DNA was isolated from peripheral blood by standard techniques and stored in the Biobank corresponding to the numbers coupled to the pedigrees described (see Figure 1a). At least 70 female hospital-based healthy control individuals from Caucasian descent were used as control DNAs.
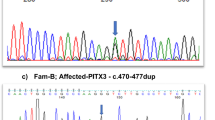
(a) Schematic representation of the NHS family pedigrees analysed in this study. Details about the full pedigree of family P8598 are described by van Dorp and Delleman.4 A (?) indicates unknown if that particular family member is a carrier, (#) indicates family member with XLC, (*) indicates family member with full manifestations of NHS), underlined pedigree numbers indicate that that particular patient received a physical exam. (b) Schematic representation of the mutations identified in the NHS gene, the mutations are listed in a similar format as previous reports describing NHS mutations, for official nomenclature of these mutations see Table 1. (Figure modified from Ramprasad et al12) (c) Photographs of the eye of a carrier female of family P8598 showing a lens opacity.
Coding exons of the NHS gene, together with splice sites were amplified using primers and the protocol described by Brooks et al.11 In this study exon 1, 2, 6, 8 were directly sequenced in both directions and exon 1b, 3, 4, 5 and 7 were analysed by dHPLC analysis according manufacturers instructions (WAVE nucleic acid fragment analysis system, Transgenomic). Fragments with shifted patterns were also sequenced in both directions.
Results
Family P 21540
The pedigree of NHS family P21540 is presented in Figure 1a. The clinical features of the probands and carriers are summarized in Table 1. One branch of this family was initially described by Pinckers et al13 as an XLC family. Unfortunately, the original family described by Pinckers refused further (follow-up) clinical examination. However, we could extend this family with another branch containing an additional patient. Patient V-1 was examined at regular intervals at our institute and had XLC, microphthalmia and abnormalities of the teeth. We identified in the NHS gene the c.1117 C>T (p.R373X) mutation. This mutation results in a premature chain termination or a truncated protein and was previously described by Burdon et al10 as a de novo mutation in a single family member with NHS.
Family P 8598
This family was initially investigated by van Dorp and Delleman.4 In summary, patients were described with X-chromosomal congenital cataract, microphthalmia, a peculiar form of the ear and dental anomalies (Table 1). The carrier females show only slight symptoms, including lens opacities (Figure 1c). Linkage analysis showed linkage of the disease locus in this pedigree to the distal Xp markers.7 Currently, we screened the entire NHS coding region, and we found in patient II:2 the c.853-2A>G (IVS3-2A>G) splice site variant. 220 healthy control alleles did not have this variant.
Also, we tested affected (I:2, II-3, II-4 and II-6) and unaffected relatives (II-1, II-5) and could only identify this mutation in affected relatives or carriers indicating that this mutation segregated with the disease phenotype. Consequently, we conclude that this sequence variant is disease causing.
Family P 20079
This family consisted of a single patient, and two probable carrier females, his mother and grandmother (Figure 1a; Table 1). Although we extended the pedigree considerably, no additional patient with NHS like features was found. The proband had congenital cataract and microphthalmia. No abnormalities of the teeth were reported, but the index patient also had apparently subnormal large ears. DNA analysis of the NHS gene showed an insertion of a single G at position 2601 on the cDNA (c.2601-2602 insG (p.K868E fsX5). This insertion disrupts the open reading frame and introduces a stop codon 14 basepairs downstream. 140 healthy control alleles did not have this variant.
Family P 24486
This family consists of a single patient, a carrier half-sister (no DNA available) and a carrier mother. The proband had congenital cataract and microcornea. He had also teeth abnormalities and large ears. Both carrier females showed congenital cataract and slight teeth abnormalities. Upon DNA analysis of the proband, a c.2635 C>T (p.R879X) mutation was found, which disrupts the NHS gene open reading frame. Examination of the DNA of the carrier mother showed the mutation in one allele. One hundred and sixty healthy control alleles did not have this variant.
Discussion
We examined four families with X-linked congenital cataract and NHS. We found protein truncation mutations and one splice site mutation causing the disease. In the families with multiple patients, considerable intrafamilial clinical variability was observed.4, 13 Clinical features ranged from isolated X-linked congenital cataract to full-blown NHS in the patients. Carrier female phenotypes ranged from no clear symptoms to cataract and mild teeth abnormalities. This is in concordance with other studies.10, 11, 12 Also in our and their studies, there is no obvious genotype/phenotype correlation between mutation position or type and severity of the disease.
Family P 8598
Here, we describe the first NHS gene splice site sequence alteration. So far, only protein truncation mutations have been described. Burdon et al10 did report, as part of a compound mutated allele, a base substitution at a splice site (IVS2-3 C>G) in combination with a frameshift mutation (c.718–719insG at the first base of exon 3) in a NHS patient. Although that base substitution was not found in 200 control alleles, the pathogenicity in NHS remained uncertain. They also identified a frameshift mutation in this patient, which resulted in a premature stop codon following the addition of 16 amino acids. This mutation is more likely to be the cause of the disease.
Interestingly, the phenotype in our pedigree of P8598 segregating with this splice site mutation appears to be more specific and less variable than observed in other large pedigrees. For instance in all affected members peculiar ear abnormalities were seen (see also van Dorp and Delleman4 for a full clinical descriptions of these abnormalities), which were apparently absent or less pronounced in other NHS pedigrees. Ramprasad et al12 observed in a large pedigree, in nearly all affected pedigree members profound ocular features but few having mild to moderate nonocular complications.
Family P 21540
The phenotypic findings in this family are quite interesting. Pinckers et al13 examined two patients from one branch of this family at a very early age, and suggested that they had XLC. Other abnormalities were not reported. That could mean that teeth abnormalities were not present at all, that they were not seen, or that development of children's teeth (milkteeth) are not, or less affected by NHS. However, a patient in another branch of the family developed full-blown NHS. The most likely explanation of our data in this family is therefore that X-linked congenital cataract and NHS are allelic disorders. Alternatively, the absence of other NHS symptoms in the young patient may either be caused by limited expression of extraocular manifestations at early age or by different effects of modifier genes between patients.
The mutation identified in this family (c.1117C>T (p.R373X)), was also described as a de novo mutation in the study of Burdon et al.10 They described that their single patient showed all typical features of NHS. There was no evidence that these families were related.
Families P 20079, P 24486
These are both families with one single patient. Both patients have a full-blown NHS phenotype and, like all other reported NHS patients so far, have a protein truncation mutation in the NHS gene. Examination of the DNA of the carrier mother of family P24486 revealed a heterozygous mutation. DNA of the carrier mother and of the grandmother of P20079 was not available. However, they were reported to have cataract, indicating that both were most likely carriers.
The mutation distribution of all pathogenic variants reported in the NHS gene so far, is shown in Figure 1b. All mutations are protein truncating, except for the one reported here in family P8598. Of the described mutations, 55% are located in exon 6. This may still represent a random distribution over the gene because exon 6 represents about 60% of the NHS gene-coding region. Alternatively or additionally, exon 6 may represent a functionally important region of the gene, since it codes for the monopartite nuclear localization signals of the NHS protein.
In conclusion, this study identified the first splice site sequence alteration mutation and three protein truncating mutations in NHS. Also, based on the differences in expression of symptoms within families, we suggest that NHS and XLC are allelic disorders.
References
Nance WE, Warburg M, Bixler D, Helveston EM : Congenital X-linked cataract, dental anomalies and brachymetacarpalia. Birth Defects Orig Art Ser 1974; 10: 285–291.
Horan MB, Billson FA : X-linked cataract and Hutchinsonian teeth. Aust Paediatr J 1974; 10: 98–102.
Toutain A, Ayrault AD, Moraine C : Mental retardation in Nance–Horan syndrome: clinical and neuropsychological assessment in four families. Am J Med Genet 1997; 22: 305–314.
van Dorp DB, Delleman JW : A family with X-chromosomal recessive congenital cataract, microphthalmia, a peculiar form of the ear and dental anomalies. J Pediatr Ophthalmol Strabismus 1979; 16: 166–171.
Walpole IR, Hockey A, Nicoll A : The Nance–Horan syndrome. J Med Genet 1990; 27: 632–634.
Zhu D, Alcorn DM, Antonarakis SE et al: Assignment of the Nance–Horan syndrome to the distal short arm of the X chromosome. Hum Genet 1990; 86: 54–58.
Bergen AA, ten Brink J, Schuurman EJ, Bleeker-Wagemakers EM : Nance–Horan syndrome: linkage analysis in a family from the Netherlands. Genomics 1994; 21: 238–240.
Toutain A, Dessay B, Ronce N et al: Refinement of the NHS locus on chromosome Xp22.13 and analysis of five candidate genes. Eur J Hum Genet 2002; 10: 516–520.
Brooks S, Ebenezer N, Poopalasundaram S et al: Refinement of the X-linked cataract locus (CXN) and gene analysis for CXN and Nance–Horan syndrome (NHS). Ophthal Genet 2004; 25: 121–131.
Burdon KP, McKay JD, Sale MM et al: Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance–Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am J Hum Genet 2003; 73: 1120–1130.
Brooks SP, Ebenezer ND, Poopalasundaram S, Lehmann OJ, Moore AT, Hardcastle AJ : Identification of the gene for Nance–Horan syndrome (NHS). J Med Genet 2004; 41: 768–771.
Ramprasad VL, Thool A, Murugan S et al: Truncating mutation in the NHS gene: phenotypic heterogeneity of Nance–Horan syndrome in an Asian Indian family. Invest Ophthalmol Vis Sci 2005; 46: 17–23.
Pinckers A, van't Pad Bosch AA, Verbeek AM, Hardus P : X-linked cataract. Ophthalm Paediatr and Genet 1982; 1: 169–172.
Acknowledgements
We thank the patients for participating in this research. We thank Professor Dr MF Niermeijer for evaluating of patients of family P20079. This project was partly sponsored by the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Florijn, R., Loves, W., Maillette de Buy Wenniger-Prick, L. et al. New mutations in the NHS gene in Nance–Horan Syndrome families from the Netherlands. Eur J Hum Genet 14, 986–990 (2006). https://doi.org/10.1038/sj.ejhg.5201671
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201671
Keywords
This article is cited by
-
A novel small deletion in the NHS gene associated with Nance-Horan syndrome
Scientific Reports (2018)
-
A novel NHS mutation causes Nance-Horan Syndrome in a Chinese family
BMC Medical Genetics (2017)
-
The first missense mutation of NHS gene in a Tunisian family with clinical features of NHS syndrome including cardiac anomaly
European Journal of Human Genetics (2011)
-
Identification of a microdeletion at Xp22.13 in a Taiwanese family presenting with Nance-Horan syndrome
Journal of Human Genetics (2011)