Abstract
In less than 10 years since its inception, RNA interference (RNAi) has had extraordinary impact on biomedical science. RNAi has been demonstrated to influence numerous biological and disease pathways. Development and adoption of RNAi technologies have been prolific ranging from basic loss-of-function tools, genome-wide screening libraries to pharmaceutical target validation and therapeutic development. However, understanding of the molecular mechanisms of RNAi is far from complete. The purpose of this brief review is to highlight key achievements in elucidating the biochemical mechanisms of the RNA-induced silencing complex and to outline major challenges for the field.
Similar content being viewed by others
Introduction
The phenomenon of RNA-induced silencing was first documented in plants by Jorgensen and co-workers 1. The field of RNA interference (RNAi) was initiated from studies conducted by Fire, Mello and co-workers 2, who induced silencing of endogenous and reporter genes in Caenorhabditis elegans following injection of long double-stranded RNA (dsRNA). Hamilton and Baulcombe 3 discovered a class of small RNA species mediating post-transcriptional silencing in plants. A seminal biochemical advance was made by Zamore et al. 4 who recapitulated dsRNA-induced silencing in vitro using Drosophila melanogaster embryo lysates. Demonstration of dsRNA-induced silencing in Drosophila S2 cells by Hannon and co-workers established a source of material amenable to biochemical purification 5. Purification of mRNA cleavage activity from dsRNA-treated S2 cells revealed a ribonucleoprotein complex comprising a nuclease and small RNA. This complex was named RNA-induced silencing complex (RISC) 5. The location at which target mRNA was cleaved corresponded to the small ∼21-23-nucleotide duplex products derived from longer dsRNA-silencing triggers 3, 6.
Small-interfering RNA
Tuschl and co-workers 7 hypothesized that these smaller RNA species directed the activity of the RISC complex. Indeed, chemically synthesized 21mer duplexes were found to trigger the cleavage of complementary mRNA in Drosophila extract 7. Perhaps the most important technological advance in the field came when Tuschl and co-workers 8 demonstrated that synthetic 21mer duplex RNA could be transfected into human cells and knockdown target expression. Further work established 21-nucleotide duplex RNA with two nucleotide 3′ overhangs as the standard small-interfering RNA (siRNA) 9. Thus, siRNA carries sequence information directing the activity of the RISC complex to specific target transcripts.
microRNA
The discovery of siRNA catalyzed our understanding of a related and previously known small RNA-mediated regulatory mechanism. The lin-4 locus encoding a 22-nucleotide non-coding RNA was identified as a temporal regulator of the lin-14 transcriptional unit 10, 11. Similarly, a 21-nucleotide let-7 RNA was found to govern the expression of a number of genes in the heterochronic lin family 12. The emergence of siRNA ushered in new methodologies for bioinformatic prediction and cloning of these endogenous microRNAs (miRNAs). It is believed that miRNAs are global regulators of gene expression involved in numerous developmental and physiological processes 13. The siRNA and miRNA pathways share many similarities including their biogenesis and silencing effects on target expression.
Dicer
The discovery of siRNA in directing RNAi intensified the search for factors that generate these small RNA species. Based on the physical properties of the siRNA duplex, Hannon and co-workers 14 deduced that members of the RNase III family were likely candidates responsible for generating siRNAs. Using expression constructs encoding epitope-tagged members of the RNase III family, analyses of immunoprecipitated material from Drosophila S2 cells revealed that long dsRNA processing was specifically attributed to an unknown protein, CG4792, which was then termed Dicer-1 14. Biochemical purification of siRNA-generating activity to homogeneity from S2 extract revealed an unknown protein CG6493, which was named Dicer-2 15. Thus, the Drosophila genome encodes two Dicer enzymes.
Analysis of the let-7 miRNA transcript predicted a possible hairpin-like secondary structure 16. Hutvagner et al. 17 demonstrated the existence of such a precursor in pupae stage Drosophila as well as in HeLa cells. Further, in vitro-transcribed let-7 precursor was processed to 22-nucleotide products in the Drosophila embryo extract. The double-stranded nature of let-7 precursor and the chemical similarities between siRNA and the let-7 miRNA raised the possibility that Dicer could generate both siRNA and miRNA. Indeed, siRNA-mediated knockdown of human Dicer in HeLa cells resulted in the attenuated production of let-7 and the accumulation of its precursor 17.
Using genetically mutated alleles in Drosophila, Lee et al. 18 identified distinct functions for Dicer-1 and Dicer-2. Dicer-1 was required for processing precursor miRNA (pre-miRNA) to miRNA, and Dicer-2 was needed for the conversion of long dsRNA to siRNA 18, 19. Recombinant Drosophila Dicer-1 was found to preferentially process pre-miRNA, and Dicer-2 preferentially processed long dsRNA 20.
In contrast, the human genome encodes only one Dicer enzyme. Hutvagner et al.17 identified the human homolog of Drosophila Dicer by using sequence homology and siRNA-mediated knockdown. Depletion of human Dicer resulted in the attenuated production of let-7 and the accumulation of its precursor 17. These findings indicated that human Dicer catalyzes the conversion of precursor miRNA to mature miRNA. Zhang et al. 21 established that recombinant human Dicer could generate both siRNA and miRNA.
Argonaute
Although Dicer was determined as the initiator nuclease of RNAi, effector components in the RISC complex remained to be identified. As in vitro RISC activity is salt sensitive, biochemical purification of the RISC complex from naïve cells is not possible (Figure 1). However, RISC complexes that are loaded with siRNA are highly resistant to salt. This can be achieved by treating cells with dsRNA. Although the biochemical basis for this marked difference is not known, (Box 1) pre-treatment of cells with dsRNA has been exploited to identify the effector machinery. The first biochemical evidence for the involvement of Argonaute (Ago) proteins in RISC came from a five-column purification of pre-loaded RISC activity to near homogeneity using long dsRNA-treated Drosophila S2 cells 22. Mass spectrometry analysis revealed a previously unidentified protein CG7439, which was named Ago2. Immunoblot analysis indicated perfect correlation between RISC activity and Ago2. Using a similar strategy, Rand et al. 23 purified pre-loaded RISC activity to near homogeneity and identified Drosophila Ago2 and siRNA as the core components of RISC.
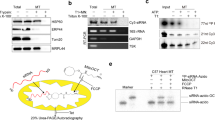
In vitro RISC activity is not recovered following anion-exchange column chromatography. Naïve Drosophila S2-S100 extract was loaded on to a HiTrap Q column and eluted with up to 1 M NaCl. Assays were performed using 100 nM duplex siRNA and 32P-labeled in vitro transcribed target mRNA and incubated at 30 °C for 30 min. Input material demonstrated high activity as most of the full-length target mRNA was cleaved. No activity is observed for flow through (FT) or bound fractions. Combining flow through and bound fractions (All) did not reconstitute activity.
Although preceding genetic studies established the involvement of Ago proteins in RNAi 24, 25, these biochemical achievements indicated a functional role for Ago. Demonstration of an Ago protein as a catalytic component of RISC came from affinity purification of Ago proteins from human cells. Affinity purification of biotinylated siRNA from HeLa cell extract identified eIF2C proteins, which represent Drosophila Ago2 homologs, as components of RISC 26. Immunoprecipitated myc-tagged mammalian Ago2 cleaved target mRNA while Ago1 and Ago3 did not 27. A similar approach using S2 cells indicated that Drosophila Ago1 and Ago2 could cleave complementary mRNA 28. Deletion and mutation analysis of epitope-tagged human Ago2 revealed specific residues comprising the catalytic site 27. This finding, in combination with crystal structures of Ago from Pyrococcus furiosus and Aquifex aeolicus, revealed a conserved aspartate-aspartate-histidine RNaseH-like active site in the PIWI domain of Ago2 29, 30, 31.
Direct demonstration of Ago2 as the slicer of RISC was achieved by the reconstitution of a “minimal RISC” 30. Recombinant human Ago2 generated from bacteria and single-strand siRNA were sufficient to catalyze the sequence-specific cleavage of target RNA. Recombinant Ago2-bearing alanine mutations in the putative active site abolished this activity. Importantly, recombinant Ago2 was not sufficient in directing duplex siRNA-mediated target cleavage 30.
dsRNA-binding proteins
RNAi is initiated by the Dicer-mediated processing of long dsRNA to siRNA duplexes. siRNA is then assembled on to the effector RISC complex to direct the sequence-specific cleavage of target mRNA. However, it was unclear as to how the initiation and effector steps were connected. A novel component of the RNAi pathway was revealed following chromatographic purification of siRNA-generating activity to homogeneity from Drosophila S2 extract 15. Mass spectrometry revealed Dicer-2 and a previously unidentified protein CG7138. This protein was named R2D2 because of its two dsRNA-binding domains (R2) and association with Dicer-2 (D2). The role of R2D2 was revealed in a partially reconstituted RISC system. Recombinant Dicer-2, Dicer-2/R2D2 or Dicer-2/mutant R2D2 were added to partially purified RISC fractions to reconstitute dsRNA- or siRNA-initiated RISC. Recombinant Dicer-2/R2D2 produced much higher RISC activity than Dicer-2 alone or Dicer-2/mutant R2D2 15. These findings were supported by genetic studies employing extract from R2D2 null flies that were fully competent for siRNA production but markedly defective for RISC activity 32. Recombinant R2D2 fully rescued this defect, possibly by bridging the initiation and effector complexes.
The discovery of a dsRNA-binding protein (dsRBP) partner for Dicer-2 raised the possibility of such a co-factor for Dicer-1. Sequence alignment and biochemical purification of miRNA-generating activity revealed a novel protein R3D1 20, 33, 34. This nomenclature derives from its three dsRNA-binding domains (R3) and its association with Dicer-1 (D1). R3D1 is also known as loquacious as mutant flies were found to de-repress the expression of a miRNA-regulated reporter gene 33, 34. Genetic mutation and RNAi knockdown of R3D1 resulted in defective pre-miRNA processing in vitro and accumulation of endogenous pre-miRNAs 20, 33, 34. Recombinant R3D1 facilitated Dicer-1-mediated miRNA production, possibly by enhancing the affinity of Dicer-1 for its substrate pre-miRNAs 20.
Domain homology search revealed two candidates as tandem partners for human Dicer. Transactivating response RNA-binding protein (TRBP) and protein activator of protein kinase R (PACT) each harbor three annotated dsRNA-binding domains and have been shown to interact with Dicer 35, 36, 37. siRNA knockdown of TRBP resulted in diminished miRNA production and loss of reporter-gene silencing in HeLa cells 35, 36. Similar results were observed in similar experiments targeting PACT 37. However, the precise role of TRBP and PACT in mediating human RISC activity remains poorly understood.
dsRBPs are not known to have any intrinsic catalytic activity. However, dsRBP-mediated enhancement of Dicer activity 20 and their requirement in partially reconstituted RISC systems 15, 32 indicate that these components are important regulators of the catalytic RNAi machinery.
RISC assembly and activation
A bi-phasic model for RISC function was now established 15, 19, 38. Dicer and a dsRBP partner generate duplex siRNA and miRNA. Ago and single-strand RNA direct the silencing of target mRNAs 22. However, it remains unknown how duplex siRNA are converted to single-strand guide RNA during RISC assembly (Box 1).
Using Drosophila embryo extracts, Nykanen et al. 39 observed the dissociation of radiolabeled duplex siRNA strands. The unwinding activity exhibited ATP dependency and eluted as a peak following gel filtration. This siRNA strand separation model was supported by observations made using biotinylated siRNA. Affinity purified human RISC complex contained only one siRNA strand 26. These findings gave rise to an unwinding model of RISC activation possibly mediated through a helicase (Figure 2A). Subsequent genetic analyses and co-immunoprecipitation studies have revealed several candidate helicases associated with known RISC components 38, 40, 41. However, no helicase has been demonstrated to function directly in the activation of RISC.
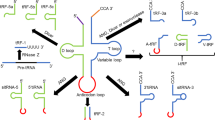
Models of RISC activation. (A) In the RISC-loading model, a dsRBP senses the more thermodynamically stable 5′ terminus of siRNA or miRNA. This sets the orientation for Dicer and dsRBP interaction with the duplex RNA. The 5′ end at which the dsRBP is bound is less preferentially loaded on to Ago. An unidentified factor separates the duplex allowing single-strand guide RNA to be loaded on to the Ago effector complex. (B) In the slicer model of RISC activation, duplex siRNA is transferred from the RISC loading machinery to Ago. Ago recognizes this configuration as the guide RNA and target RNA, cleaving the passenger strand.
siRNA incorporation into RISC can be asymmetric 42. That is, one siRNA strand is preferentially loaded on to RISC. Computational analyses revealed that miRNA from human, mouse, fly and worm exhibited a defined thermodynamic signature 43. The 5′ and median nucleotides of the incorporated miRNA strand had lower thermodynamic stability relative to other positions. This profile was also observed for functional but not non-functional siRNA. A direct test for these rules was made by rescuing non-functional siRNA through introducing 5′ and internal mismatches 42, 43. These findings indicate that RISC assembly is an asymmetric process.
Tomari et al. 44 initiated studies aimed at elucidating the interaction of Dicer-2 and R2D2 with siRNA. Crosslinking experiments indicated a possible role for R2D2 in sensing thermodynamic siRNA structure. R2D2 marked the more stable 5′ terminus excluding this siRNA strand from RISC. Subsequent studies revealed that this potential asymmetric sensing mechanism was possible for mature siRNA and for siRNA processed from long dsRNA precursors 45. Using radiolabeled siRNA and native gel electrophoresis, Drosophila Dicer-2 and R2D2 were identified in an RISC loading complex required for the formation of RISC 19, 44. These studies outline a role for Dicer and dsRBP in generating, loading and activating RISC in an asymmetric manner.
Slicer-mediated RISC activation
The slicer function of Ago has been proposed as an alternative mechanism in the process of RISC assembly. In contrast to siRNA strand separation models, the slicer model of RISC activation outlines the transfer of duplex siRNA from the RISC loading machinery to Ago2 46, 47. Duplex RNA loaded on Drosophila Ago2 is then acted upon as the guide and the target resulting in cleavage of the passenger strand (Figure 2B). Using radiolabeled passenger strand annealed to unlabeled guide strand, the predicted labeled 9mer passenger strand cleavage product was detected in Drosophila embryo lysates 28, 46, 47. Further, no 9mer cleavage products were identified in extracts from ago2 null embryos 47, nor from r2d2 or dicer-2 null embryos 46. These experiments indicate the possibility of slicer-mediated activation of RISC.
However, the generality of this mechanism of RISC activation is unclear. To date, among the four human Agos, cleavage activity has only been demonstrated for human Ago2, yet transfected siRNA has been shown to interact with all four human Ago proteins 48. Moreover, duplexes bearing mismatches such as that of miRNA do not undergo passenger strand cleavage 46. Thus, a cleavage model of RISC activation may not be all encompassing. It is possible that there are multiple mechanisms of RISC activation including those mediated through siRNA strand displacement and passenger strand cleavage. Clearly, the biochemical events that occur between the action of Dicer and formation of the active RISC complex are poorly understood and represent a long-standing challenge for the field (Box 1).
RISC effector mechanisms
The best understood mechanism by which RISC executes RNAi is Ago-mediated cleavage of target mRNA. Following siRNA administration to mice and using an RNA ligase and cDNA amplification protocol, Soutschek et al. 49 demonstrated the recovery of target mRNA cleaved at predicted sites.
One of the enduring dogmas in the field is the distinction between siRNA-induced RISC (siRISC) and that induced by miRNA (miRISC). Identification of two Dicers in Drosophila and their preferences for different substrates support the possibility of parallel pathways 18, 20. Also, Drosophila Ago2 is required for siRISC and Ago1 is required for miRISC 50. It has been suggested that guide RNA fully complementary to target mRNA induces siRISC-mediated cleavage and partial complementarity results in miRISC-mediated translational repression 51, 52.
It is worth clarifying that the net effect of RNAi is inhibition of target mRNA expression. Mechanisms by which this occurs can be classified as those that are dependent and those that are independent of Ago-mediated cleavage. However, the distinction between siRISC and miRISC may not be as simple as the parallel model predicts: (1) many species have only one Dicer; (2) human Dicer and Drosophila Dicer-1 can process both long dsRNA and pre-miRNA 20, 21; (3) although they are often distinguished by the degree of inter-strand complementarity, siRNA and miRNA share similar secondary structure, and algorithms used to design effective synthetic siRNAs are based on thermodynamic profiles of miRNAs 42, 43; (4) transfected siRNAs have been shown to load on to all four human Ago proteins 48; (5) siRNAs have been shown to cleave target mRNAs that are less than fully complementary 53, and reporter constructs bearing imperfect let-7 target sites were found to be cleaved in human cells 54; (6) human miRNA-196 was found to cleave HOXB8 mRNA, an endogenous target transcript 55; (7) there is increasing evidence that miRNA reduces target mRNA levels 56, 57. Thus, the degree of distinction and crosstalk among various RNAi pathways is not currently known. If RNAi is viewed as a genomic regulatory mechanism, then overlap of pathways and mechanistic redundancy would aid in functionality.
Understanding the cleavage-independent mechanisms of RNAi represent a large gap in the field (Box 1). A long-standing association between RNAi components and translational machinery suggests a connection between these two processes 58. Co-immunoprecipitation and co-fractionation of polysomes with target mRNA and RISC components are most often used in such studies 59. Various antibiotics and cap-dependent and cap-independent expression constructs have been employed to study miRISC-mediated inhibition of translation initiation 52. A piwi protein was recently shown to interact with the mRNA cap-binding complex 60. Further, the association between a factor involved in regulating mRNA stability and miRISC activity further indicate potential crosstalk between these cellular functions 61. Coupling of RNAi with active translation would be an efficient means by which the RISC complex could survey transcriptome activity. However, as a minimal RISC complex, containing single-strand guide RNA and recombinant Ago is sufficient to effect target cleavage 30; the importance of protein synthesis in the recognition of mRNA by RISC is unclear (Box 1). Indeed, in vitro assays that mimic mechanisms of RISC function independent of transcript cleavage would be tremendously useful for the field.
A cellular mechanism for RISC has also emerged from findings of processing (P) bodies 62. P-bodies are cytoplasmic foci containing RISC components and mRNA decapping and deadenylation factors 63, 64, 65. A strong association has been shown for miRNA, target mRNA expression and P-body markers in human cells 61. Stress granules are additional cellular structures that accumulate Ago proteins in a miRNA-dependent manner 66. Support for these localization-mediated mechanisms of RNAi would be enhanced by elucidating how miRNA and their targets approximate in these regions and identifying factors that mediate these interactions.
Perspective
The central problem in understanding the biochemical mechanisms of RISC is an incomplete knowledge of the factors involved. Although genetic approaches have implicated a number of candidate factors, the precise role of these in effecting RNAi is unknown (Box 1). To date, Dicer, dsRBPs and Ago are the only RISC components that have been validated biochemically. Affinity-purified complexes from human cell extracts containing these three components can induce RISC activity in vitro 67, 68. However, recombinant Dicer, dsRBPs and Ago are not sufficient to reconstitute RISC (Liu Q, unpublished).
Creative and imaginative assays amenable to biochemical purification need to be developed to further advance the field (Box 1). The need for this is even more pressing in light of the new classes of small regulatory RNAs including piwi-interacting RNA (piRNA) 69, 70 and repeat-associated siRNA (rasiRNA) 71. An example of the potential usefulness of alternative assays was demonstrated by Kingston and co-workers 72 who purified end-labeled piRNA from rat testis. Following five chromatograph column separations, piwi and RecQ1 were identified as a candidate effector complex for this class of small regulatory RNA 72. As with siRISC and miRISC, elucidating biochemical mechanisms of small RNA biogenesis, complex assembly and function for these new pathways represent future challenges for the field.
Reconstituting the holo-RISC enzyme would be a benchmark achievement. That is, defining the RNAi machinery such that the activation and effector mechanisms of RISC are revealed would bring much needed mechanistic insight. Beyond this, however, understanding regulatory mechanisms that govern RNAi would connect the field to the wider biological context (Box 1). In principle, biochemical regulation of RNAi has been established 73. The influence of cellular signaling pathways on RISC activity and the contribution of RNAi to physiological processes are critical in understanding the importance of small regulatory RNAs in biology and disease.
References
Napoli C, Lemieux C, Jorgensen R . Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990; 2:279–289.
Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806–811.
Hamilton AJ, Baulcombe DC . A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999; 286:950–952.
Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA . Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 1999; 13:3191–3197.
Hammond SM, Bernstein E, Beach D, Hannon GJ . An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000; 404:293–296.
Zamore PD, Tuschl T, Sharp PA, Bartel DP . RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000; 101:25–33.
Elbashir SM, Lendeckel W, Tuschl T . RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001; 15:188–200.
Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494–498.
Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T . Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 2001; 20:6877–6888.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–854.
Wightman B, Ha I, Ruvkun G . Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75:855–862.
Reinhart BJ, Slack FJ, Basson M et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403:901–906.
He L, Hannon GJ . MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522–531.
Bernstein E, Caudy AA, Hammond SM, Hannon GJ . Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409:363–366.
Liu Q, Rand TA, Kalidas S, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003; 301:1921–1925.
Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408:86–89.
Hutvagner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834–838.
Lee YS, Nakahara K, Pham JW, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004; 117:69–81.
Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ . A Dicer-2-dependent 80 s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 2004; 117:83–94.
Jiang F, Ye X, Liu X, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 2005; 19:1674–1679.
Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W . Single processing center models for human Dicer and bacterial RNase III. Cell 2004; 118:57–68.
Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ . Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001; 293:1146–1150.
Rand TA, Ginalski K, Grishin NV, Wang X . Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci USA 2004; 101:14385–14389.
Tabara H, Sarkissian M, Kelly WG, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 1999; 99:123–132.
Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H . AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA 2000; 97:11650–11654.
Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T . Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002; 110:563–574.
Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004; 305:1437–1441.
Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC . Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 2005; 19:2837–2848.
Song JJ, Smith SK, Hannon GJ, Joshua-Tor L . Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004; 305:1434–1437.
Rivas FV, Tolia NH, Song JJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 2005; 12:340–349.
Yuan YR, Pei Y, Ma JB, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 2005; 19:405–419.
Liu X, Jiang F, Kalidas S, Smith D, Liu Q . Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 2006; 12:1514–1520.
Forstemann K, Tomari Y, Du T, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 2005; 3:e236.
Saito K, Ishizuka A, Siomi H, Siomi MC . Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 2005; 3:e235.
Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005; 436:740–744.
Haase AD, Jaskiewicz L, Zhang H, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 2005; 6:961–967.
Lee Y, Hur I, Park SY, et al. The role of PACT in the RNA silencing pathway. EMBO J 2006; 25:522–532.
Tomari Y, Du T, Haley B, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 2004; 116:831–841.
Nykanen A, Haley B, Zamore PD . ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001; 107:309–321.
Meister G, Tuschl T . Mechanisms of gene silencing by double-stranded RNA. Nature 2004; 431:343–349.
Meister G, Landthaler M, Peters L, et al. Identification of novel argonaute-associated proteins. Curr Biol 2005; 15:2149–2155.
Schwarz DS, Hutvagner G, Du T, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003; 115:199–208.
Khvorova A, Reynolds A, Jayasena SD . Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115:209–216.
Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD . A protein sensor for siRNA asymmetry. Science 2004; 306:1377–1380.
Preall JB, He Z, Gorra JM, Sontheimer EJ . Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr Biol 2006; 16:530–535.
Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD . Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005; 123:607–620.
Rand TA, Petersen S, Du F, Wang X . Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005; 123:621–629.
Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 2004; 15:185–197.
Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004; 432:173–178.
Okamura K, Ishizuka A, Siomi H, Siomi MC . Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 2004; 18:1655–1666.
Hutvagner G, Zamore PD . A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002; 297:2056–2060.
Petersen CP, Bordeleau ME, Pelletier J, Sharp PA . Short RNAs repress translation after initiation in mammalian cells. Mol Cell 2006; 21:533–542.
Haley B, Zamore PD . Kinetic analysis of the RNAi enzyme complex. Nat Struc Mol Biol 2004; 11:599–606.
Schmitter D, Filkowski J, Sewer A, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic acids Res 2006; 34:4801–4815.
Yekta S, Shih IH, Bartel DP . MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004; 304:594–596.
Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433:769–773.
Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005; 122:553–563.
Ishizuka A, Siomi MC, Siomi H . A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 2002; 16:2497–2508.
Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005; 309:1573–1576.
Grivna ST, Pyhtila B, Lin H . MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA 2006; 103:13415–13420.
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W . Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006; 125:1111–1124.
Sheth U, Parker R . Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003; 300:805–808.
Liu J, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 2005; 7:1261–1266.
Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R . MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 2005; 7:719–723.
Behm-Ansmant I, Rehwinkel J, Doerks T, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885–1898.
Leung AKL, Calabrese JM, Sharp PA . Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA 2006; 103:18125–18130.
Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R . Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005; 123:631–640.
Maniataki E, Mourelatos Z . A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 2005; 19:2979–2990.
Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006; 442:203–207.
Girard A, Sachidanandam R, Hannon GJ, Carmell MA . A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006; 442:199–202.
Vagin VV, Sigova A, Li C, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320–324.
Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science 2006; 313:363–367.
Kennedy S, Wang D, Ruvkun G . A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 2004; 427:645–649.
Acknowledgements
Xiaodong Wang is an investigator with the Howard Hughes Medical Institute. Qinghua Liu is a W.A. “Tex” Moncrief Jr Scholar in Medical Research and a Damon Runyon Scholar supported by the Damon Runyon Cancer Research Foundation (DRS-43). This work was also supported by a Welch grant (I-1608) and an NIH grant (GM078163).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Paroo, Z., Liu, Q. & Wang, X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Res 17, 187–194 (2007). https://doi.org/10.1038/sj.cr.7310148
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310148
Keywords
This article is cited by
-
An unnatural enzyme with endonuclease activity towards small non-coding RNAs
Nature Communications (2023)
-
Epigenetic interaction of microbes with their mammalian hosts
Journal of Biosciences (2021)
-
Effects of silencing key genes in the capsanthin biosynthetic pathway on fruit color of detached pepper fruits
BMC Plant Biology (2014)
-
Efficient inhibition of HIV-1 replication by an artificial polycistronic miRNA construct
Virology Journal (2012)
-
ABC transporters and RNAi in Caenorhabditis elegans
Journal of Bioenergetics and Biomembranes (2007)