Abstract
EKLF is an erythroid-specific, zinc finger-containing transcription factor essential for the activation of the mammalian beta globin gene in erythroid cells of definitive lineage. We have prepared a polyclonal anti-mouse EKLF antibody suitable for Western blotting and immunoprecipitation (IP) qualities, and used it to define the expression patterns of the EKLF protein during mouse erythroid development. We have also used this antibody for the chromatin-immunoprecipitation (ChIP) assay. EKLF was found to bind in vivo at both the mouse beta-major-globin promoter and the HS2 site of beta-LCR in the mouse erythroleukemia cells (MEL) in a DMSO-inducible manner. The DMSO-induced bindings of EKLF as well as three other proteins, namely, RNA polymerase II, acetylated histone H3, and methylated histone H3, were not abolished but significantly lowered in CB3, a MEL-derived cell line with null-expression of p45/NF-E2, an erythroid-enriched factor needed for activation of the mammalian globin loci. Interestingly, binding of EKLF in vivo was also detected in the mouse alpha-like globin locus, at the adult alpha globin promoter and its far upstream regulatory element alpha-MRE (HS26). This study provides direct evidence for EKLF-binding in vivo at the major regulatory elements of the mouse beta-like globin gene clusters the data also have interesting implications with respect to the role of EKLF-chromatin interaction in mammalian globin gene regulation.
Similar content being viewed by others
Introduction
Expression of the eukaryotic globin genes in the erythroid cells is regulated in a tissue- and developmental stage-specific manner 1, 2, 3, 4. The murine β-like globin gene family consists of four functional gene members arranged and expressed during development in the order of 5′-εy-βH1-βmaj-βmin-3′ 5. In the embryonic yolk sac, higher levels of εy and βH1 are expressed. On day 11.5 of gestation, the main site for erythropoiesis changes from yolk sac to the fetal liver. Concomitantly, the globin expression is switched from εy and βH1 to the βmaj and βmin forms 6. Similar arrangement of the β-like globin genes and their developmental regulation (switch) are also observed in humans 1, 2, 3, 4. The regulation of the mammalian β-like globin switch is achieved through the assembly and interactions of multiple DNA-protein and protein-protein complexes at the locus-control-region (β-LCR) and different globin upstream promoter regions 1, 2, 3, 4. Among the various nuclear factors participating in the regulation of the mammalian globin switch are those expressed in an erythroid-specific or erythroid-enriched manner, including NF-E2 7, GATA-1 8, FOG 9 and EKLF.
EKLF is a transcription factor exclusively expressed in the erythroid tissues 10, 11. In EKLF-knockout mice, the embryonic erythropoiesis and expression of the embryonic εy gene is normal. However, they died of anemia before E16 due to failure to express the adult β globin 12, 13. In interesting contrast, overexpressing EKLF in mice induced an earlier switch from the fetal to adult-type globin 14. These studies have demonstrated that EKLF is essential for activation of the adult β globin gene. Analysis of transgenic mice carrying human β-like globin gene cluster suggested a similar requirement of EKLF for human adult β globin gene activation 15, 16, 17.
Mechanistically, the function of EKLF is carried out distinctively and separately by its proline-rich transcription activation domain at the N-terminus and the three C2H2-type zinc fingers at the C-terminus 18, 19, 20, 21. Through these zinc fingers, EKLF binds the CACCC box in vitro in the β globin promoter region. Furthermore, intactness of the CACCC box is required for EKLF mediated β globin gene activation in vivo 22. Interestingly, the zinc fingers of EKLF have been found to carry out a wide range of functions aside from DNA-binding. Nuclear targeting of EKLF is mediated by its zinc fingers through interaction in vivo with the import in proteins 23, 24. The zinc finger region of EKLF is also the site for interaction with the SWI/SNF complex. This interaction resulted in the remodeling of the chromatin and the formation of the DNA hypersensitive site (HS) at the β globin promoter in vitro 20. A protein complex, E-RCl that could activate transcription of a chromatin-assembled β globin promoter in an EKLF-dependent fashion has been purified 25. Consistent with the in vitro results, lack of EKLF expression led to the loss of the HS at the β globin promoter in vivo 12, 13. The above studies together have pointed to a model of activation of the mammalian adult β globin gene by EKLF. That is, EKLF binds the β globin promoter through the interaction between its zinc fingers and CACCC box in the promoter. This interaction attracts chromatin remodeling and transcription activating complexes to the β globin promoter.
Due to the lack of appropriate antibodies, several interesting questions about the interaction between EKLF and its target chromatin regions remained unanswered. For example, does EKLF indeed bind in vivo at the globin promoter of the erythroid cells? Also, the CACCC box is present in other regions of the β-like globin locus, including the HS of the β-LCR. In particular, the HS3 site of the human β-LCR appears to be required for EKLF-dependent activation of a cis-linked human β globin promoter in transgenic mice 14. Thus, does EKLF bind in erythroid cells at one or more of the HS of β-LCR as well? In the following, we report the use of an affinity-purified anti-EKLF antibody for ChIP analysis of EKLF binding in vivo in the murine β-like as well as the α-like globin gene clusters of the murine adult erythroleukemia cell line MEL. Both α-like and β-like globin genes are silent in uninduced MEL, but their transcription activities are greatly turned-on upon treatment with certain inducers such as dimethyl sulfoxide (DMSO) 26. It was found that EKLF indeed binds in the chromatin region of the β globin promoter. Interestingly, EKLF binding in vivo was also easily detected at HS2, but not HS3, of the β-LCR, as well as at the α MRE (H26), the LCR/enhancer of the murine α-like globin locus. Finally, we have found that the levels of EKLF binding at these chromatin regions in CB3, a MEL derivative lacking the expression of p45/NF-E2, were all significantly lower than those detected in MEL.
Materials and methods
Plasmid construction and DNA transfection
For induction of MEL and CB3, cells at a density of 2×105/ml were added 1.5% DMSO and the culturing was continued for 96 hours. HA-tagged EKLF was PCR amplified from the reverse transcription products of the induced MEL RNA, and cloned into the Hind III and Not I sites of pCMV vector (Invitrogen). The cDNA sequence corresponds to aa 1–376 of mouse EKLF. DNA transfection of 293 cells was performed by the calcium phosphate-DNA coprecipitation method. 2 μg of the pCMV-HA-EKLF plasmid was transfected into 5×105 cells.
Cell culture
The murine adult erythroleukemia cell line MEL 27, human 293, and human HeLa cells were all cultured in DMEM medium (Invitrogen) containing 10% FBS (Hyclone), 50 unit/ml of penicillin, and 50 μg/ml of streptomycin (Invitrogen). CB3, a MEL derivative lacking the expression of p45 28, was grown in RPMI 1640 medium (Invitrogen) containing the same ingredients as above. The procedures for the establishment of CB3 pools stably expressing human p45/NF-E2 followed those described in ref 29.
Antibodies
The anti-p45/NF-E2 rabbit antibody C-19 and anti-RNA polymerase II (anti-pol II) rabbit antibody N-20 were purchased from Santa Cruz Inc. The anti-tubulin, anti-HA and anti-actin mouse antibodies were from Sigma. The anti-acetyl-histone H3 (anti-AcH3) and anti-dimethyl-lysine 4 of histone H3 (H3MeK4) antibodies were from Upstate. Polyclonal antibody recognizing mouse EKLF was generated in the rabbit by using tandemly arranged, trimeric repeats of the peptide sequence SEETQDLGPG (aa 51 to 60 of EKLF) as the antigen, as described in 30. Affinity-purified antibody was prepared by passing 2 ml of the rabbit anti sera through a gel column packed with 0.15 g of CNBr-activated Sepharose 4B (Amersham Pharmacia Biotech) coupled with the SEETQDLGPG peptide. Antibodies retained on the column were eluted with six column volumes (3 ml) of the elution buffer (0.1 M glycine, pH 2.0) and then neutralized with 1 M Tris buffer, pH 9.0. The concentration of the antibody was measured with the Bio-Rad protein assay kit. Due to the relatively low amount of the purified antibody, the eluate had to be concentrated to 2.2 μg/ml for later uses. The antibody was stored at −20 °C before use in Western blotting, IP and ChIP analysis.
Extract preparation
The whole cell extracts were prepared from MEL and CB3 cells by lysis of the cell pellets containing 5×106 of PBS-washed cells in 300 ml of M-PER® Reagent (PIERCE). To prepare extracts from different mouse tissues, 2 month old B6 mice were sacrificed, and the cells were lysed and homogenized in T-PER® Reagent (PIERCE) with the ratio of 0.1 g tissue per 200 μl of the reagent.
Western blotting analysis
For Western blot assays, aliquots (20 μg) of the whole cell extracts were electrophoresed on 10% sodium dodecyl sulfate/10%-polyacrylamide gel (SDS-PAGE), and electrotransferred to a nitrocellulose membrane (Amersham Biosciences). Staining with PonceauS (Pierce) was used to estimate the loading and blotting efficiencies. The blots were hybridized with the primary and then the secondary antibodies prepared in Tris-buffered saline solution containing 5% dry milk. The hybridizing bands were identified by the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences).
Northern blot analysis
The Northern blot hybridization procedures essentially followed those of Liu et al. 31. The total RNAs were extracted by means of the commercial Trizol reagent (Invitrogen), loaded on 1% agarose gels containing 6% formaldehyde, and transferred to a nylon membrane. After hybridization with radioactive DNA probes amplified by PCR, the blots were subjected to autoradiography.
Immunoprecipitation (IP)
The IP experiments were done as follows. The cells were lysed in the Mammalian Protein Extraction Reagent (M-PER Pierce) containing the protease inhibitor cocktail from Roch, 1 mM PMSF, 60 μg/ml MG132 (Calbiochen), and 20 μg/ml NEM (Sigma). The lysates were incubated with the anti-EKLF or anti-HA antibody (Sigma) for 4 h, added the protein A and G Sepharose beads (Pharmacia), and further incubation was carried out for another 3 h. The beads were then spun down, washed three times with the lysis buffer, boiled in the SDS sample buffer, and finally loaded on the SDS-PAGE gel. The gel was then blotted and hybridized with anti-EKLF or anti-HA as described above.
Chromatin immunoprecipitation (ChIP)
The ChIP analysis followed the procedures by Daftari et al.32. 2×106 confluent MEL cells with or without treatment with 1.5% DMSO for 96 h were fixed at room temperature for 10 min in 60 ml of DMEM medium containing 1% formaldehyde. After sonication, the protein-DNA complexes were immunoprecipitated with anti-EKLF, anti-p45, anti-H3MeK4, anti-AcH3 or anti-pol II. The precipitated chromatin DNAs were purified and amplified by PCR reactions using the appropriate primers: 95°C for 7 min, followed by 34 cycles at 94°C for 30 s, 55°C for 40 s, and 72°C for 40 s. The primers used in PCR of different mouse genomic regions and the lengths of the PCR fragments generated are as follows: HS3 (283 bp), 5′-GAG GAG AAA ACA AAC AGG AGG TCT-3′ and 5′-CCT TTG TCC TAC TGC TCT CAT GTT-3′; HS2 (277 bp), 5′-CAC TTC TTC ATA TTC TCT CTC TAG-3′ and 5′-CTT ATT TTC TTT TCA CCT TCC CTG-3′; βmaj promoter regions (217 bp), 5′-GAC AAA CAT TAT TCA GAG GGA GTA-3′ and 5′-AAG CAA ATG TGA GGA GCA ACT GA T-3′ 33; α MRE (HS26) (311 bp), 5′-ACC CAT CTG GAA CCT ATG AG-3′ and 5′-GAA AGT CTT CCC AAC TGC AG-3′; α1 promoter region (243 bp), 5′-CTT CCC AAA CTG CCA TCA CT-3′ and 5′-TCT TGG ACC TGC AGC AAG TA-3′, ζ promoter region (234 bp) 5′-AAT GAC CTC CTG ACA TGG CT-3′ and 5′-TAT ACA CAT GGT CAG GGA CC-3′. Those for the two intergenic regions are α intergenic (520 bp), 5′-AGA ACC TTC AAG GCG GAT GCT-3′ and 5′-GTC CCA GAC AAG TAA GTG GCT-3′; β-intergenic (271 bp), 5′-TCC TGG TTT ATG AGC GAG AAG AA-3′ and 5′-AGA CCT GAT CCC CAT TCC TAT TCC-3′ 33. The DNA amounts used for PCR were determined by the intensities of the β-actin signals. The PCR signals were first normalized to those from the β-actin region. The different target/ β-actin ratios were then further normalized against the target/ β-actin ratios of the input samples, and used to plot the histographs. The relative intensities of the positive signals were given as the folds of increase over those from the preimmune samples. Each histogram consists of averages of data derived from 2-3 sets of PCR analysis conducted with the use of chromatin DNAs precipitated from at least two different MEL pools. The folds of differences are given as mean±SEM.
Results
Analysis of the levels of EKLF protein during development with an affinity purified polyclonal antibody (AEK-1)
For the purpose of analysis of EKLF expression and its targets of binding in vivo in erythroid cells, we have produced a rabbit anti-mouse EKLF antibody, AEK-1, using the approach by Hsu et al. 30. This method takes advantage of the fact that tandem repeats of an epitope sequence often possess potent antigenicity 30.
Previously, the expression of EKLF in different cell lines and tissues has been analyzed only by Northern blotting 10, 11, 34. In those studies, it was found that the human EKLF RNA was detectable in bone marrow cells and the erythroleukemia cell lines HEL, JK1 and OCIM1, but not in the erythroid K562 nor in the myeloid or lymphoid cell lines 11. Mouse EKLF RNA was expressed in the adult bone marrow, spleen, MEL and mouse bone marrow-derived cell lines 10. To examine the tissue-specificity and stage-specificity of the expression of the EKLF protein, we analyzed by Western blotting the extracts prepared from the murine embryo yolk sac (E 9.5 day), different tissues from E 14.5 day fetal mice, and different tissues of 2 month old mice.
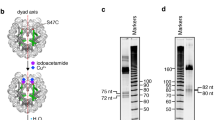
As shown in Figure 1A, use of AEK-1 as the probe for Western blotting detected a single band of the approximate molecular weight 40 kDa in MEL cell extract (lane 1, Figure 1A), which is absent in the 293 extract (lane 2, Figure 1A). It could also recognize the tagged HA-EKLF expressed in transfected 293 cells (lane 3, Figure 1A). The IP property of AEK-1 was demonstrated in Figure 1B, in which AEK-1 could specifically pull down EKLF from MEL cell extract (lane 1 of the left panel, Figure 1B), but not from those of the non-erythroid HeLa and 293 cells (lanes 2 and 4 of the left panel, Figure 1B). The antigenecity of AEK-1 was supported by IP experiments of the transfected 293 cells (middle and right panels, Figure 1B).
Western blotting analysis of mouse EKLF. (A) Western blotting property of AEK-1. The anti-mouse EKLF rabbit antibody AEK-1 was used to probe Western blot containing whole cell extracts prepared from: lane 1, MEL cells; lane 2, 293T cells; lane 3, 293T cells transfected with pHA-EKLF. As control, the blot was also probed with anti-tubulin. The positions of HA-EKLF and EKLF are indicated with the open and black arrowheads, respectively. (B) IP property of AEK-1. Left panel-Whole cell extracts prepared from MEL (lane 1), HeLa (lane 2), and 293T (lane 4) cells were immunoprecipitated with AEK-1 and then analysed by Western blotting as probed with AEK-1. The IP of MEL was also carried out with preimmune serum (PI, lane 3). The black arrowhead points to the expected gel position of EKLF. Middle and right panels-Whole cell extracts prepared from 293T cells transfected with pEF-HA-EKLF were immunoprecipitated with anti-HA (middle panel) and AEK-1(right panel), and then analyzed by Western blotting as probed with the anti-HA antibody. (C) Expression patterns of EKLF in different mouse tissues. Whole cell extracts were prepared from different mouse tissues and analyzed by Western blotting with AEK-1 as the probe. The open arrowhead points to the gel position of a non-specific band on the blot. Note the presence of EKLF in the extracts of yolk sac, fetal liver and adult spleen.
As seen in Figure 1C, the EKLF protein is expressed in the embryonic yolk sac, the fetal liver, and the adult spleen, all of which are the mouse erythroid tissues. Due to the heterogeneity of erythroid cell populations in the different tissues, it is difficult to have an accurate quantitative comparison. However, Western blot analysis, with actin as the control, showed that the level of EKLF protein is highest in the adult spleen, followed by the fetal liver, and then the yolk sac. This is similar to the changes of the EKLF mRNA levels during development, as observed by Southwood et al. 34.
Analysis of in vivo binding of EKLF in the murine β-like globin gene cluster of MEL
Based on the IP quality of the antibody AEK-1, we have used it to investigate EKLF-binding in vivo in the β-like globin locus as analyzed by the mammalian ChIP method 32. This approach has been used to analyze factor-chromatin interaction or the mammalian β-like 32, 33, 35, 36, 37, 38, 39, 40, 41 and α-like globin loci 32, 42, 43 of different erythroid cell lines and tissues.
To do this, we fixed MEL cells with or without DMSO induction by formaldehyde before sonication, and immunoprecipitated the fixed chromatin fragments with AEK-1 and with antibodies directed against p45/NF-E2, histone H3 methylated at K4 (H3MeK4), acetylated histone H3 (AcH3), and RNA polymerase II (pol II). as described in the Materials and methods.
The precipitated DNAs were then analyzed by PCR for the extents of enrichment of different genomic region, as shown in the panels of Figure 2A and 2B.
Chromatin immunoprecipitation assay (ChIP) of the mouse β-like globin gene locus. ChIP was used to analyze the association in vivo of different protein factors with HS2, HS3, and βmaj promoter of the mouse β-like globin locus. The map of the globin locus is shown on top of (A). DNA precipitated from MEL (A and B) and CB3 (C) cells, with (+) or without (−) DMSO induction, with the use of different antibodies and preimmune serum (PI) were analyzed by PCR using primers specific for HS2 and HS3 of β-LCR, βmaj promoter, an intergenic region, and β-actin gene, respectively. The PCR products were then analyzed by agarose gel electrophoresis. The DNA amounts used for PCR were determined by the intensities of the β-actin signals. The PCR signals were first normalized to those from the β-actin region. The different target/β-actin ratios were then further normalized against the target/β-actin ratios of the input samples, and used to plot the histographs. The relative intensities of the positive signals were given as the folds of increase over those from the preimmune samples. Each histogram consists of averages of data derived from 2-3 sets of PCR analysis conducted with the use of chromatin DNAs precipitated from three different cell pools. The folds of differences are given as mean ± SEM. Negative controls of the ChIP samples were prepared with the use of the preimmune serum. The relative intensities of the positive signals were given as the folds of increase over those from the preimmune samples.
Specific binding of p45/NF-E2, H3MeK4, AcH3 and pol II at HS2 as well as the βmaj globin promoter could be easily detected in uninduced MEL, but the levels were not very high (-DMSO, Figure 2A). For example, the relative enrichments of these factors at HS2 were 3.3-, 1.4-, 1.8, and 1.4-fold, respectively. The binding of these proteins were all elevated in DMSO-induced MEL, in which the βmaj globin gene became transcriptionally active (+DMSO, blank bars, Figure 2A). For example, the relative enrichments of binding of the four factors at HS2 became 9-, 6-, 4-, and 2-fold, respectively, in DMSO-treated MEL (black bars, Figure 2A). The data are consistent with the previous studies 32, 33, 35, 36, 37, 38, 39, 40, 41, although we have observed an induction, instead of reduction, of the levels of H3MeK4 binding at both the βmaj promoter and HS2 in DMSO treated MEL.
More interestingly, use of AEK-1 has demonstrated that EKLF indeed binds at the βmaj promoter region in DMSO-induced MEL (Figure 2A). This is consistent with our earlier genomic-footprinting analysis, in which it was shown that the promoter was empty in uninduced MEL, and it became occupied with factor(s) upon DMSO induction 44. On the other hand, no apparent enrichment of EKLF binding in the εy and βh1 globin promoter could be detected (data not shown). Interestingly, induced EKLF-binding could also be detected at HS2 of the β-LCR (Figure 2A). In contrast to HS2, there was little, if any, binding of EKLF at HS3 site of the β-LCR (Figure 2B). The above ChIP data suggested that EKLF could directly bind in vivo at both the βmaj promoter and HS2 of β-LCR. Alternatively, this might be the result of the close proximity of the two regulatory elements during looping of the chromosomal domain of the β-like globin gene cluster (1–4).
Reduced EKLF binding in the β-like globin locus in the p45-null cell line CB3
CB3 is a derivative of MEL in which the p45 expression is blocked due to a viral integration event 28. To see whether there is a functional link between p45 and EKLF with respect to factor-chromatin interaction, we have carried out ChIP analysis of CB3.
As expected, no p45-binding could be detected at all of the genome regions assayed, including HS2, the βmaj promoter, an intergenic region of the β-like globin locus, and the β-actin gene (Figure 2C). On the other hand, the patterns and inducibility of binding of EKLF, pol II, H3MeK4, and AcH3 at HS2 were all similar to those observed in MEL (compare Figures 2C to 2A). In interesting contrast to HS2, the levels of induced binding of the above four proteins at the βmaj promoter all decreased significantly in CB3 as compared to MEL (compare the histographs of Figure 2C and 2A). Consistent with the previous studies 35, 39, 40, 41, this result indicated that null-expression of p45 in CB3 had a greater effect on the chromatin structure and factor-binding in the βmaj promoter region than those in HS2 of the β-LCR. Our data also suggested that if EKLF was involved in HS3 mediated activation of the β globin gene promoter 14, it might not do so through binding at HS3.
EKLF binding in the murine α-like globin locus
We have also used ChIP to analyze the chromatin structure and factor-binding of the murine α-like globin locus in MEL as well as in CB3. As shown in Figure 3A, we observed enriched and DMSO-induced binding of p45, pol II, H3MeK4, and AcH3 at the α globin promoter and, to a less extent, αMRE (HS26), which is the upstream enhancer/LCR sequence of the murine α-like globin locus. The binding patterns of these four proteins are in general agreement with the previous studies 42, 43. Interestingly but also surprisingly, DMSO-induced binding of EKLF was detected at the HS26 as well as the a promoter (Figure 3A), the regulation of which has never been linked to EKLF (see discussions later). In contrast, there was no apparent enrichment of EKLF binding at either the embryonic z globin promoter or the intergenic region of the murine a locus (Figure 3A).
ChIP analysis of the mouse a-like globin locus. ChIP analysis was preformed as described in Figure 2, except that PCRs of the precipitated chromatin DNAs were carried out with the use of primers specific for the α-MRE (H 26), the α promoter, the ζ promoter, and an intergenic region of the mouse α-like globin locus, the map of which is shown on top of (A). Again, both the MEL (A) and CB3 (B) cells were analyzed.
Similar to the β locus, a significant effect of null-expression of p45 on the chromatin structure and factor-binding of the α-like globin locus was also observed as well. The inducibilities of binding of all five proteins including EKLF were significantly lower in CB3 than in MEL (compare Figure 3B to 3A).
Reduced expression of EKLF in CB3 cells
The reduced binding of EKLF at the two murine globin loci in CB3 than in MEL could be the result of inefficient assembly of EKLF into multiple protein-DNA complexes at the LCRs and the promoters due to the lack of p45, which also binds in vivo to a set of regulatory elements that EKLF binds to (Figure 2 and 3). Alternatively, the reduced binding might be simply because of a reduced level of EKLF protein in CB3 than in MEL.
To partially answer this, we have analyzed, by Northern and Western blotting, the levels of EKLF in CB3 and MEL. As shown in Figure 4A, both the mRNA and protein levels of EKLF in CB3 are approximately 50% that of MEL. This data suggest that at least the lower amount of EKLF in CB3 has contributed to the reduced binding of this factor in the regulatory regions of the two globin loci in this cell line.
(A) Comparison of EKLF expression levels of MEL and CB3. The expression levels of EKLF in MEL and CB3, with (+) or without (−) DMSO induction, were analyzed by Northern blotting and Western blotting analyses. As the control, the expression levels of G3PDH and tubulin were also analyzed. The non-erythroid 293T sample was included for the purpose of comparison. (B) Failure of rescue of EKLF expression by p45. CB3 cell pools overexpressing human p45 were established, and the levels of p45, EKLF and tubulin were analyzed by Western blotting. Note that the level of EKLF was not significantly increased, as exemplified for one of the CB3 (p45) pools, by the overexpression of human p45.
It is not known, however, whether and how p45 might regulate the expression of EKLF in MEL cells. It is worthy to mention here that overexpression of human p45 in CB3 could not rescue the expression level of EKLF (Figure 4B).
Discussion
Unlike the other erythroid-enriched transcription factors including NF-E2 and GATA-1, report on the status of EKLF-binding in the regulatory regions of the β-like globin locus has been lacking, possibly due to the availability of suitable antibodies. In this study, we have used a newly acquired anti-EKLF antibody, AEK-1, to directly study the physical interaction between EKLF and the globin gene regulatory regions in erythroid cells by the mammalian ChIP method 32. Our results have provided further support to some of the previous structural analysis of the chromatin domain of the β-like globin locus. More importantly, the data also shed new light on the functional aspects of EKLF in globin gene regulation.
First, consistent with the previous genomic footprinting analysis 44, while little, if any, EKLF binding could be detected in the βmaj promoter in MEL cells, there was already some, although not much, EKLF-binding in HS2. Upon DMSO induction, both regions became highly enriched in EKLF (Figure 2A). This result is in striking similarity to the change of p45/NF-E2 binding at the βmaj promoter and HS2 during MEL induction 33, 36, 37. In particular, Groudine and his colleagues have suggested that before DMSO induction of MEL, HS2 was bound predominantly with sMafK/Bach 1 heterodimer although some NF-E2 molecules, which are heterodimers of p45 and sMafK, could also be detected at the site 36, 45. Upon DMSO induction, it was mainly NF-E2 binding at HS2. Concomitant with this, the level of binding of p45/NF-E2 at the βmaj promoter was also significantly enhanced 33, 36, 37. We suggest that during MEL differentiation, EKLF and NF-E2 may be coregulated through similar pathway(s) with respect to their physical movements to the β-like globin locus and the consequent activation of the adult βmaj globin gene. The exact reason for why EKLF binds more efficiently to the globin loci in DMSO-induced MEL cells remains unknown at the moment. It could be due to cooperative interaction among EKLF and other factors including NF-E2. Alternatively, differences in the post-translational modification and/or subcellular location of EKLF, as induced by DMSO, may play a role. Further experiments, including the use of an antibody suitable for immunostaining, are needed to solve this interesting question.
In relation to the above, Tewari R, et al. have provided evidence that EKLF function in mice might also be regulated through HS3 besides HS2 14. However, we did not detect EKLF binding at the HS3 site in the DMSO-induced MEL cells. There are several possible reasons for this. For one thing , MEL is a cell line. Secondly, HS3 of MEL cells could still have EKLF binding, but in the context of a complex of multiple factors. The EKLF epitope recognizable by our antibody might be masked by these other factors in the complex.
Second, we have found that EKLF was also bound at the regulatory region H26 and the adult α, but not embryonic ζ, globin promoter in MEL as well as in CB3 (Figure 3). This is somewhat surprising because EKLF (−/−) mice exhibited little change in their a globin expression. This paradox may be similar to that previously documented for p45. This factor, as a subunit of the heterodimeric NF-E2, is a transcription activator of two globin loci 7 and, yet, p45 (−/−) mice showed no defect in globin gene regulation 46. We suggest that, similar to the scenario of “functional degeneracy” as proposed for the above observation of p45 (−/−) mice, EKLF is involved in the regulation of both the β- and α-like globin loci in mammals including mice. When both EKLF alleles were deleted, the βmaj globin expression was blocked because of the lack of EKLF binding at β-LCR and the βmaj promoter. However, in the α-like globin locus, another factor(s) could move in and take over the physical as well as functional positions of EKLF, and thus maintain the developmental expression of the α-like globin locus in the EKLF (−/−) mice.
Finally, with the availability of AEK-1, it is now possible to analyze the developmental stage specificities of EKLF binding in the two globin loci, as well as the sub-cellular distributions of this factor in the different erythroid tissues during development.
References
Bulger M, Groudine M . Looping versus linking: toward a model for long-distance gene activation. Genes Dev 1999; 13:2465–2477.
Engel JD, Tanimoto K . Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 2000; 100:499–502.
Li Q, Peterson KR, Fang X, Stamatoyannopoulos G . Locus control regions. Blood 2002; 100:3077–3086.
de Laat W, Grosveld F . Spatial organization of gene expression: the active chromatin hub. Chromosome Res 2003; 11:447–459.
Kingsley PD, Malik J, Emerson RL, et al. “Maturational” globin switching in primary primitive erythroid cells. Blood 2006; 107:1665–1672.
Whitney JB, 3rd . Differential control of the synthesis of two hemoglobin beta chains in normal mice. Cell 1977; 12:863–871.
Andrews NC . The NF-E2 transcription factor. Int J Biochem Cell Biol 1998; 30:429–432.
Martin DI, Tsai SF, Orkin SH . Increased gamma-globin expression in a nondeletion HPFH mediated by an erythroid-specific DNA-binding factor. Nature 1989; 338:435–438.
Tsang AP, Visvader JE, Turner CA, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 1997; 90:109–119.
Miller IJ, Bieker JJ . A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol 1993; 13:2776–2786.
Bieker JJ . Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol 1996; 15:347–352.
Perkins AC, Sharpe AH, Orkin SH . Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 1995; 375:318–322.
Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F . Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 1995; 375:316–318.
Tewari R, Gillemans N, Wijgerde M, et al. Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5'HS3 of the beta-globin locus control region. Embo J 1998; 17:2334–2341.
Gillemans N, Tewari R, Lindeboom F, et al. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev 1998; 12:2863–2873.
Tanimoto K, Liu Q, Grosveld F, Bungert J, Engel JD . Context-dependent EKLF responsiveness defines the developmental specificity of the human epsilon-globin gene in erythroid cells of YAC transgenic mice. Genes Dev 2000; 14:2778–2794.
Wijgerde M, Gribnau J, Trimborn T, et al. The role of EKLF in human beta-globin gene competition. Genes Dev 1996; 10:2894–2902.
Bieker JJ, Southwood CM . The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol Cell Biol 1995; 15:852–860.
Chen X, Bieker JJ . Unanticipated repression function linked to erythroid Kruppel-like factor. Mol Cell Biol 2001; 21:3118–3125.
Kadam S, McAlpine GS, Phelan ML, et al. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev 2000; 14:2441–2451.
Pandya K, Donze D, Townes TM . Novel transactivation domain in erythroid Kruppel-like factor (EKLF). J Biol Chem 2001; 276:8239–8243.
Bieker JJ . Kruppel-like factors: three fingers in many pies. J Biol Chem 2001; 276:34355–34358.
Pandya K, Townes TM . Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J Biol Chem 2002; 277:16304–16312.
Quadrini KJ, Bieker JJ . Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of erythroid Kruppel-like factor. J Biol Chem 2002; 277:32243–32252.
Armstrong JA, Bieker JJ, Emerson BM . A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 1998; 95:93–104.
Geller R, Levenson R, Housman D . Significance of the cell cycle in commitment of murine erythroleukemia cells to erythroid differentiation. J Cell Physiol 1978; 95:213–222.
Nudel U, Salmon JE, Terada M, et al. Differential effects of chemical inducers on expression of beta globin genes in murine erythroleukemia cells. Proc Natl Acad Sci USA 1977; 74:1100–1104.
Lu SJ, Rowan S, Bani MR, Ben-David Y . Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci USA 1994; 91:8398–8402.
Shyu YC, Lee TL, Ting CY, et al. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol Cell Biol 2005; 25:10365–10378.
Hsu CT, Ting CY, Ting CJ, et al. Vaccination against gonadotropin-releasing hormone (GnRH) using toxin receptor-binding domain-conjugated GnRH repeats. Cancer Res 2000; 60:3701–3705.
Liu JJ, Hou SC, Shen C-KJ . Erythroid gene suppression by NF-kappa B. J Biol Chem 2003; 278:19534–19540.
Daftari P, Gavva NR, Shen C-KJ . Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene 1999; 18:5482–5486.
Sawado T, Igarashi K, Groudine M . Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci USA 2001; 98:10226–10231.
Southwood CM, Downs KM, Bieker JJ . Erythroid Kruppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev Dyn 1996; 206:248–259.
Kiekhaefer CM, Grass JA, Johnson KD, Boyer ME, Bresnick EH . Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc Natl Acad Sci USA 2002; 99:14309–14314.
Brand M, Ranish JA, Kummer NT, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 2004; 11:73–80.
Sawado T, Halow J, Bender MA, Groudine M . The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 2003; 17:1009–1018.
Forsberg EC, Downs KM, Christensen HM, et al. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci USA 2000; 97:14494–14499.
Johnson KD, Grass JA, Park C, et al. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol 2003; 23:6484–6493.
Forsberg EC, Downs KM, Bresnick EH . Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood 2000; 96:334–339.
Johnson KD, Christensen HM, Zhao B, Bresnick EH . Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell 2001; 8:465–471.
Anguita E, Johnson CA, Wood WG, Turner BM, Higgs DR . Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc Natl Acad Sci USA 2001; 98:12114–12119.
Anguita E, Hughes J, Heyworth C, et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. Embo J 2004; 23:2841–2852.
Reddy PM, Shen C-KJ . Erythroid differentiation of mouse erythroleukemia cells results in reorganization of protein-DNA complexes in the mouse beta maj globin promoter but not its distal enhancer. Mol Cell Biol 1993; 13:1093–1103.
Francastel C, Magis W, Groudine M . Nuclear relocation of a transactivator subunit precedes target gene activation. Proc Natl Acad Sci USA 2001; 98:12120–12125.
Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, et al. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 1995; 81:695–704.
Acknowledgements
We thank our lab colleagues for various reagents and for their help. We also appreciate the discussions on the topic with Jim Bieker. This research was supported by a grant from the National Health Research Institute (NHRI), and by the Academia Sinica, Taipei.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shyu, YC., Wen, SC., Lee, TL. et al. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res 16, 347–355 (2006). https://doi.org/10.1038/sj.cr.7310045
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310045
Keywords
This article is cited by
-
An extra-erythrocyte role of haemoglobin body in chondrocyte hypoxia adaption
Nature (2023)
-
Direct competition between DNA binding factors highlights the role of Krüppel-like Factor 1 in the erythroid/megakaryocyte switch
Scientific Reports (2017)
-
miR-122-mediated translational repression of PEG10 and its suppression in human hepatocellular carcinoma
Journal of Translational Medicine (2016)
-
Differential regulation of the α-globin locus by Krüppel-like factor 3 in erythroid and non-erythroid cells
BMC Molecular Biology (2014)
-
Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells
Nature Genetics (2010)