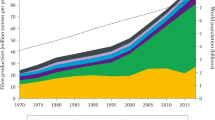
Abstract
The demand for sustainable and environmentally friendly sources of foods and cosmetics is increasing owing to concerns about the rapid growth of the global population and climate change. Microorganisms are promising cell factories in which to produce various food and cosmetic ingredients. However, the commercialization of microbial-based food and cosmetic compounds is still limited by the insufficient performances of microbial strains and processes. Systems metabolic engineering can improve the performance of microorganisms. In this Review, we highlight food and cosmetic compounds that can be produced using microorganisms. We then discuss systems metabolic engineering, with its different phases that include production mode and host selection, metabolic pathway reconstruction, tolerance enhancement, metabolic flux and fermentation process optimization, downstream process integration and scale-up, which can be optimized to improve the development of high-performance microbial processes. Finally, we overview the current limitations and future directions of industrializing microbial processes for the production of food and cosmetic compounds.
Key points
-
Microorganisms can serve as green factories for the production of food and cosmetic products.
-
A growing number of food and cosmetic compounds are commercially produced from renewable substrates using microorganisms.
-
Systems metabolic engineering could facilitate the development of high-performance microbial cell factories.
-
Various tools and strategies are powering up systems metabolic engineering, such as synthetic/systems biology tools and process/evolutionary engineering strategies.
-
Raw materials, performance of microbial strains, fermentation and downstream processes, process scale-up, economic aspects, and societal demands and perception should be collectively considered for the successful commercialization of microbial processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Choi, K. R., Yu, H. E. & Lee, S. Y. Microbial food: microorganisms repurposed for our food. Microb. Biotechnol. 15, 18–25 (2022).
Behnassi, M. & El Haiba, M. Implications of the Russia–Ukraine war for global food security. Nat. Hum. Behav. 6, 754–755 (2022).
Choi, K. R. et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837 (2019). This article reports the tools and strategies of systems metabolic engineering and recent trends.
Ko, Y. S. et al. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 49, 4615–4636 (2020).
Park, J. H., Lee, K. H., Kim, T. Y. & Lee, S. Y. Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl Acad. Sci. USA 104, 7797–7802 (2007). This article was the first to report the application of systems metabolic engineering to improve the microbial production of compounds.
Lee, K. H., Park, J. H., Kim, T. Y., Kim, H. U. & Lee, S. Y. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol. Syst. Biol. 3, 149 (2007).
Zhang, D. et al. Reducing lactate secretion by ldhA deletion in l-glutamate- producing strain Corynebacterium glutamicum GDK-9. Braz. J. Microbiol. 45, 1477–1483 (2014).
Ahn, J. H. et al. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat. Commun. 11, 1970 (2020).
Huang, J. et al. High yield and cost‐effective production of poly (γ‐glutamic acid) with Bacillus subtilis. Eng. Life Sci. 11, 291–297 (2011).
Jiang, L. et al. Aeration and fermentation strategies on nisin production. Biotechnol. Lett. 37, 2039–2045 (2015).
Zhu, Y. et al. Construction of allitol synthesis pathway by multi-enzyme coexpression in Escherichia coli and its application in allitol production. J. Ind. Microbiol. Biotechnol. 42, 661–669 (2015).
Patel, S. N., Kaushal, G. & Singh, S. P. A novel d-allulose 3-epimerase gene from the metagenome of a thermal aquatic habitat and d-allulose production by Bacillus subtilis whole-cell catalysis. Appl. Environ. Microbiol. 86, e02605-19 (2020).
Shin, K. C., Sim, D. H., Seo, M. J. & Oh, D. K. Increased production of food-grade d-tagatose from d-galactose by permeabilized and immobilized cells of Corynebacterium glutamicum, a GRAS host, expressing d-galactose isomerase from Geobacillus thermodenitrificans. J. Agric. Food Chem. 64, 8146–8153 (2016).
Jagtap, S. S. & Rao, C. V. Production of d-arabitol from d-xylose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Appl. Microbiol. Biotechnol. 102, 143–151 (2018).
Li, S. et al. Dynamic control over feedback regulatory mechanisms improves NADPH flux and xylitol biosynthesis in engineered E. coli. Metab. Eng. 64, 26–40 (2021).
Yuan, X. et al. Increasing NADPH availability for xylitol production via pentose-phosphate-pathway gene overexpression and Embden–Meyerhof–Parnas-pathway gene deletion in Escherichia coli. J. Agric. Food Chem. 69, 9625–9631 (2021).
Li, M. H. et al. Enhanced production of dihydroxyacetone from glycerol by overexpression of glycerol dehydrogenase in an alcohol dehydrogenase-deficient mutant of Gluconobacter oxydans. Bioresour. Technol. 101, 8294–8299 (2010).
Yang, L. B. et al. A novel osmotic pressure control fed-batch fermentation strategy for improvement of erythritol production by Yarrowia lipolytica from glycerol. Bioresour. Technol. 151, 120–127 (2014).
Yang, J. et al. Pathway construction in Corynebacterium glutamicum and strain engineering to produce rare sugars from glycerol. J. Agric. Food Chem. 64, 9497–9505 (2016).
Liu, J. J. et al. l-Fucose production by engineered Escherichia coli. Biotechnol. Bioeng. 116, 904–911 (2019).
Ni, Z. et al. Multi-path optimization for efficient production of 2’-fucosyllactose in an engineered Escherichia coli C41 (DE3) derivative. Front. Bioeng. Biotechnol. 8, 611900 (2020).
Wang, Y. et al. Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum. Nat. Commun. 11, 3120 (2020).
Cerminati, S. et al. Low cost and sustainable hyaluronic acid production in a manufacturing platform based on Bacillus subtilis 3NA strain. Appl. Microbiol. Biotechnol. 105, 3075–3086 (2021).
Jin, P., Kang, Z., Yuan, P., Du, G. & Chen, J. Production of specific-molecular-weight hyaluronan by metabolically engineered Bacillus subtilis 168. Metab. Eng. 35, 21–30 (2016).
Zhou, Z. et al. A microbial-enzymatic strategy for producing chondroitin sulfate glycosaminoglycans. Biotechnol. Bioeng. 115, 1561–1570 (2018).
Kim, H. M., Chae, T. U., Choi, S. Y., Kim, W. J. & Lee, S. Y. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nat. Chem. Biol. 15, 721–729 (2019).
Yu, T. et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558.e14 (2018).
Okuda, T. et al. Omega-3 eicosatetraenoic acid production by molecular breeding of the mutant strain S14 derived from Mortierella alpina 1S-4. J. Biosci. Bioeng. 120, 299–304 (2015).
Okuda, T. et al. Eicosapentaenoic acid (EPA) production by an oleaginous fungus Mortierella alpina expressing heterologous the Δ17‐desaturase gene under ordinary temperature. Eur. J. Lipid Sci. Technol. 117, 1919–1927 (2015).
Li, Z. et al. Overexpression of malonyl-CoA: ACP transacylase in Schizochytrium sp. to improve polyunsaturated fatty acid production. J. Agric. Food Chem. 66, 5382–5391 (2018).
Guo, D. S., Ji, X. J., Ren, L. J., Li, G. L. & Huang, H. Improving docosahexaenoic acid production by Schizochytrium sp. using a newly designed high‐oxygen‐supply bioreactor. AIChE J. 63, 4278–4286 (2017).
Nyyssola, A., Suhonen, A., Ritala, A. & Oksman-Caldentey, K. M. The role of single cell protein in cellular agriculture. Curr. Opin. Biotechnol. 75, 102686 (2022).
Banks, M., Johnson, R., Giver, L., Bryant, G. & Guo, M. Industrial production of microbial protein products. Curr. Opin. Biotechnol. 75, 102707 (2022).
Bilal, M. et al. Bioprospecting and biotechnological insights into sweet-tasting proteins by microbial hosts — a review. Bioengineered 13, 9815–9828 (2022).
Raveendran, S. et al. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 56, 16–30 (2018).
Okpara, M. O. Microbial enzymes and their applications in food industry: a mini-review. Adv. Enzyme Res. 10, 23–47 (2022).
Gupta, P. L., Rajput, M., Oza, T., Trivedi, U. & Sanghvi, G. Eminence of microbial products in cosmetic industry. Nat. Prod. Bioprospect. 9, 267–278 (2019).
Sunar, K., Kumar, U. & Deshmukh, S. in Agro-Industrial Wastes as Feedstock for Enzyme Production (eds Dhillon, G. S. & Kaur, S.) 279–298 (Elsevier, 2016).
Chen, H. et al. A stepwise control strategy for glutathione synthesis in Saccharomyces cerevisiae based on oxidative stress and energy metabolism. World J. Microbiol. Biotechnol. 36, 117 (2020).
Kongklom, N., Shi, Z., Chisti, Y. & Sirisansaneeyakul, S. Enhanced production of poly-γ-glutamic acid by Bacillus licheniformis TISTR 1010 with environmental controls. Appl. Biochem. Biotechnol. 182, 990–999 (2017).
Zhou, L., Deng, C., Cui, W. J., Liu, Z. M. & Zhou, Z. M. Efficient l-alanine production by a thermo-regulated switch in Escherichia coli. Appl. Biochem. Biotechnol. 178, 324–337 (2016).
Yamamoto, S. et al. Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 78, 4447–4457 (2012).
Hasegawa, S. et al. Improvement of the redox balance increases l-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl. Environ. Microbiol. 78, 865–875 (2012).
Huang, Q., Liang, L., Wu, W., Wu, S. & Huang, J. Metabolic engineering of Corynebacterium glutamicum to enhance l-leucine production. Afr. J. Biotechnol. 16, 1048–1060 (2017).
Yin, L., Zhao, J., Chen, C., Hu, X. & Wang, X. Enhancing the carbon flux and NADPH supply to increase l-isoleucine production in Corynebacterium glutamicum. Biotechnol. Bioprocess. Eng. 19, 132–142 (2014).
Xu, J. Z., Ruan, H. Z., Yu, H. B., Liu, L. M. & Zhang, W. Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of l-lysine production from mixed sugar. Microb. Cell Fact. 19, 39 (2020).
Zhang, X. et al. High-yield production of l-serine through a novel identified exporter combined with synthetic pathway in Corynebacterium glutamicum. Microb. Cell Fact. 19, 115 (2020).
Wang, C., Wu, J., Shi, B., Shi, J. & Zhao, Z. Improving l-serine formation by Escherichia coli by reduced uptake of produced l-serine. Microb. Cell Fact. 19, 66 (2020).
Liu, Y. et al. Genetic engineering of Escherichia coli to improve l-phenylalanine production. BMC Biotechnol. 18, 5 (2018).
Patnaik, R., Zolandz, R. R., Green, D. A. & Kraynie, D. F. l-Tyrosine production by recombinant Escherichia coli: fermentation optimization and recovery. Biotechnol. Bioeng. 99, 741–752 (2008). This article reports a systematically optimized scaled-up fermentation for tyrosine production and the downstream processes for recovering and purifying tyrosine.
Kim, B., Binkley, R., Kim, H. U. & Lee, S. Y. Metabolic engineering of Escherichia coli for the enhanced production of l-tyrosine. Biotechnol. Bioeng. 115, 2554–2564 (2018).
Du, L., Zhang, Z., Xu, Q. & Chen, N. Central metabolic pathway modification to improve l-tryptophan production in Escherichia coli. Bioengineered 10, 59–70 (2019).
Wu, H. et al. Highly efficient production of l-histidine from glucose by metabolically engineered Escherichia coli. ACS Synth. Biol. 9, 1813–1822 (2020).
Yang, D. et al. Expanded synthetic small regulatory RNA expression platforms for rapid and multiplex gene expression knockdown. Metab. Eng. 54, 180–190 (2019).
Zhao, L. et al. Expression regulation of multiple key genes to improve l-threonine in Escherichia coli. Microb. Cell Fact. 19, 46 (2020).
Huang, J. F. et al. Systematic analysis of bottlenecks in a multibranched and multilevel regulated pathway: the molecular fundamentals of l-methionine biosynthesis in Escherichia coli. ACS Synth. Biol. 7, 2577–2589 (2018).
Li, B. et al. Rerouting fluxes of the central carbon metabolism and relieving mechanism-based inactivation of l-aspartate-α-decarboxylase for fermentative production of β-alanine in Escherichia coli. ACS Synth. Biol. 11, 1908–1918 (2022).
Man, Z. et al. Systems pathway engineering of Corynebacterium crenatum for improved l-arginine production. Sci. Rep. 6, 28629 (2016).
Park, S. H. et al. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat. Commun. 5, 4618 (2014). This article reports SysME strategies for developing industrially competitive Corynebacterium glutamicum strain for l-arginine production.
Xu, M. et al. Development of a novel biosensor-driven mutation and selection system via in situ growth of Corynebacterium crenatum for the production of l-arginine. Front. Bioeng. Biotechnol. 8, 175 (2020).
Zhang, J. et al. De novo engineering of Corynebacterium glutamicum for l-proline production. ACS Synth. Biol. 9, 1897–1906 (2020).
Hao, N. et al. Improvement of l-citrulline production in Corynebacterium glutamicum by ornithine acetyltransferase. J. Ind. Microbiol. Biotechnol. 42, 307–313 (2015).
Zhang, B., Gao, G., Chu, X. H. & Ye, B. C. Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of l-ornithine driven by glucose and xylose. Bioresour. Technol. 284, 204–213 (2019).
Liu, H., Hou, Y., Wang, Y. & Li, Z. Enhancement of sulfur conversion rate in the production of l-cysteine by engineered Escherichia coli. J. Agric. Food Chem. 68, 250–257 (2020).
Bang, H. B., Lee, Y. H., Kim, S. C., Sung, C. K. & Jeong, K. J. Metabolic engineering of Escherichia coli for the production of cinnamaldehyde. Microb. Cell Fact. 15, 16 (2016).
Luo, Z. W. & Lee, S. Y. Metabolic engineering of Escherichia coli for the production of benzoic acid from glucose. Metab. Eng. 62, 298–311 (2020).
Guo, J. et al. Cloning and identification of a novel tyrosinase and its overexpression in Streptomyces kathirae SC-1 for enhancing melanin production. FEMS Microbiol. Lett. 362, fnv041 (2015).
Chavez-Bejar, M. I. et al. Metabolic engineering of Escherichia coli to optimize melanin synthesis from glucose. Microb. Cell Fact. 12, 108 (2013).
Grewal, P. S., Modavi, C., Russ, Z. N., Harris, N. C. & Dueber, J. E. Bioproduction of a betalain color palette in Saccharomyces cerevisiae. Metab. Eng. 45, 180–188 (2018).
Zhou, S., Hao, T. & Zhou, J. Fermentation and metabolic pathway optimization to de novo synthesize (2S)-naringenin in Escherichia coli. J. Microbiol. Biotechnol. 30, 1574–1582 (2020).
Du, Y., Yang, B., Yi, Z., Hu, L. & Li, M. Engineering Saccharomyces cerevisiae coculture platform for the production of flavonoids. J. Agric. Food Chem. 68, 2146–2154 (2020).
Eichenberger, M., Hansson, A., Fischer, D., Durr, L. & Naesby, M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 18, foy046 (2018).
Yang, J. et al. Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis. Metab. Eng. 59, 44–52 (2020).
Jones, J. A. et al. Complete biosynthesis of anthocyanins using E. coli polycultures. mBio 8, e00621-17 (2017).
Hansen, E. H. et al. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 75, 2765–2774 (2009).
Luo, Z. W., Cho, J. S. & Lee, S. Y. Microbial production of methyl anthranilate, a grape flavor compound. Proc. Natl Acad. Sci. USA 116, 10749–10756 (2019).
Dinh, C. V. & Prather, K. L. J. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc. Natl Acad. Sci. USA 116, 25562–25568 (2019).
Du, J., Yang, D., Luo, Z. W. & Lee, S. Y. Metabolic engineering of Escherichia coli for the production of indirubin from glucose. J. Biotechnol. 267, 19–28 (2018).
Yang, D., Park, S. Y. & Lee, S. Y. Production of rainbow colorants by metabolically engineered Escherichia coli. Adv. Sci. 8, e2100743 (2021).
Choi, K. R., Yu, H. E., Lee, H. & Lee, S. Y. Improved production of heme using metabolically engineered Escherichia coli. Biotechnol. Bioeng. 119, 3178–3193 (2022). This article reports strategies for optimizing a fermentation process for production of haem using an engineered Escherichia coli strain.
Ko, Y. J. et al. Animal-free heme production for artificial meat in Corynebacterium glutamicum via systems metabolic and membrane engineering. Metab. Eng. 66, 217–228 (2021).
Choi, K. R., Yu, H. E. & Lee, S. Y. Production of zinc protoporphyrin IX by metabolically engineered Escherichia coli. Biotechnol. Bioeng. 119, 3319–3325 (2022).
Ghiffary, M. R. et al. High-level production of the natural blue pigment indigoidine from metabolically engineered Corynebacterium glutamicum for sustainable fabric dyes. ACS Sustain. Chem. Eng. 9, 6613–6622 (2021).
Giesselmann, G. et al. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnol. J. 14, e1800417 (2019).
Liu, W. et al. Engineering Escherichia coli for high-yield geraniol production with biotransformation of geranyl acetate to geraniol under fed-batch culture. Biotechnol. Biofuels 9, 58 (2016).
Wang, X. et al. Engineering Escherichia coli for production of geraniol by systematic synthetic biology approaches and laboratory-evolved fusion tags. Metab. Eng. 66, 60–67 (2021).
Dusseaux, S., Wajn, W. T., Liu, Y., Ignea, C. & Kampranis, S. C. Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids. Proc. Natl Acad. Sci. USA 117, 31789–31799 (2020).
Rolf, J., Julsing, M. K., Rosenthal, K. & Lutz, S. A gram-scale limonene production process with engineered Escherichia coli. Molecules 25, 1881 (2020).
Liu, Y. et al. Engineering the oleaginous yeast Yarrowia lipolytica for production of alpha-farnesene. Biotechnol. Biofuels 12, 296 (2019).
Meadows, A. L. et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537, 694–697 (2016). This article reports a systematically developed industrially relevant microbial strain for β-farnesene production.
Cha, Y. et al. Probing the synergistic ratio of P450/CPR to improve (+)-nootkatone production in Saccharomyces cerevisiae. J. Agric. Food Chem. 70, 815–825 (2022).
Mikkelsen, M. D. et al. Methods for improved production of rebaudioside D and rebaudioside M. US patent 10,612,066 (2014).
Zhu, Z. T. et al. Metabolic compartmentalization in yeast mitochondria: burden and solution for squalene overproduction. Metab. Eng. 68, 232–245 (2021).
Li, D. et al. Metabolic engineering of Yarrowia lipolytica for heterologous oleanolic acid production. Chem. Eng. Sci. 218, 115529 (2020).
Sun, Z. J. et al. Combined biosynthetic pathway engineering and storage pool expansion for high-level production of ergosterol in industrial Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 9, 681666 (2021).
Guo, X. J. et al. Compartmentalized reconstitution of post-squalene pathway for 7-dehydrocholesterol overproduction in Saccharomyces cerevisiae. Front. Microbiol. 12, 663973 (2021).
Sun, T. et al. Production of lycopene by metabolically-engineered Escherichia coli. Biotechnol. Lett. 36, 1515–1522 (2014).
Luo, Z. et al. Enhancing isoprenoid synthesis in Yarrowia lipolytica by expressing the isopentenol utilization pathway and modulating intracellular hydrophobicity. Metab. Eng. 61, 344–351 (2020).
Larroude, M. et al. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of beta-carotene. Biotechnol. Bioeng. 115, 464–472 (2018).
Shen, H. J. et al. Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab. Eng. 38, 180–190 (2016).
Gong, Z. et al. Coordinated expression of astaxanthin biosynthesis genes for improved astaxanthin production in Escherichia coli. J. Agric. Food Chem. 68, 14917–14927 (2020).
Park, S. Y., Eun, H., Lee, M. H. & Lee, S. Y. Metabolic engineering of Escherichia coli with electron channelling for the production of natural products. Nat. Catal. 5, 726–737 (2022). This study reports the electron channelling strategy to improve the microbial production of lutein and other natural products.
Sun, L., Kwak, S. & Jin, Y. S. Vitamin A production by engineered Saccharomyces cerevisiae from sylose via two-phase in situ extraction. ACS Synth. Biol. 8, 2131–2140 (2019).
Han, M. & Lee, P. C. Microbial production of bioactive retinoic acid using metabolically engineered Escherichia coli. Microorganisms 9, 1520 (2021).
Hu, Q., Zhang, T., Yu, H. & Ye, L. Selective biosynthesis of retinol in S. cerevisiae. Bioresour. Bioprocess. 9, 22 (2022).
van Maris, A. J. et al. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl. Environ. Microbiol. 70, 159–166 (2004).
Forster, A., Aurich, A., Mauersberger, S. & Barth, G. Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 75, 1409–1417 (2007).
Li, C., Gao, S., Yang, X. & Lin, C. S. K. Green and sustainable succinic acid production from crude glycerol by engineered Yarrowia lipolytica via agricultural residue based in situ fibrous bed bioreactor. Bioresour. Technol. 249, 612–619 (2018).
Chung, S. C., Park, J. S., Yun, J. & Park, J. H. Improvement of succinate production by release of end-product inhibition in Corynebacterium glutamicum. Metab. Eng. 40, 157–164 (2017).
Yamane, T. & Tanaka, R. Highly accumulative production of l(+)-lactate from glucose by crystallization fermentation with immobilized Rhizopus oryzae. J. Biosci. Bioeng. 115, 90–95 (2013).
Brock, S., Kuenz, A. & Prüße, U. Impact of hydrolysis methods on the utilization of agricultural residues as nutrient source for d-lactic acid production by Sporolactobacillus inulinus. Fermentation 5, 12 (2019).
Tsuge, Y. et al. Metabolic engineering of Corynebacterium glutamicum for hyperproduction of polymer-grade l- and d-lactic acid. Appl. Microbiol. Biotechnol. 103, 3381–3391 (2019).
Jiang, L. et al. Enhanced butyric acid tolerance and bioproduction by Clostridium tyrobutyricum immobilized in a fibrous bed bioreactor. Biotechnol. Bioeng. 108, 31–40 (2011).
Deng, Y. et al. Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab. Eng. 46, 28–34 (2018).
Pereira, B. et al. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab. Eng. 34, 80–87 (2016).
Liu, M., Ding, Y., Xian, M. & Zhao, G. Metabolic engineering of a xylose pathway for biotechnological production of glycolate in Escherichia coli. Microb. Cell Fact. 17, 51 (2018).
Bae, S. J., Kim, S. & Hahn, J. S. Efficient production of acetoin in Saccharomyces cerevisiae by disruption of 2,3-butanediol dehydrogenase and expression of NADH oxidase. Sci. Rep. 6, 27667 (2016).
Li, Z. et al. Systems metabolic engineering of Corynebacterium glutamicum for high-level production of 1,3-propanediol from glucose and xylose. Metab. Eng. 70, 79–88 (2022).
Yamamoto, S., Suda, M., Niimi, S., Inui, M. & Yukawa, H. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol. Bioeng. 110, 2938–2948 (2013).
Su, H., Chen, H. & Lin, J. Enriching the production of 2-methyl-1-butanol in fermentation process using Corynebacterium crenatum. Curr. Microbiol. 77, 1699–1706 (2020).
Zhao, E. M. et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687 (2018).
Xin, F., Basu, A., Yang, K. L. & He, J. Strategies for production of butanol and butyl-butyrate through lipase-catalyzed esterification. Bioresour. Technol. 202, 214–219 (2016).
Yang, D., Jang, W. D. & Lee, S. Y. Production of carminic acid by metabolically engineered Escherichia coli. J. Am. Chem. Soc. 143, 5364–5377 (2021). This study reports the microbial production of carminic acid by applying SysME strategies.
Kim, H.-S. et al. Microorganism producing 5′-inosinic acid and process for producing 5′-inosinic acid using the same. US patent 7,244,608 (2007).
Cho, J., Kim, H. W., Oh, Y. S. & Park, J. H. Corynebacteria strain for enhancement of 5′-guanosine monophosphate productivity and a method of producing 5′-guanosine monophosphate using the same. US patent 8,530,200 (2013).
Lee, K. H. et al. Microorganism for producing riboflavin and method for producing riboflavin using the same. US patent 7,166,456 (2007).
Serrano-Amatriain, C. et al. Folic acid production by engineered Ashbya gossypii. Metab. Eng. 38, 473–482 (2016).
Maekawa, T. & Zhang, Z. Y. Method for producing vitamin B12 from hydrogen-metabolizing methane bacterium. US patent 7,018,815 (2006).
Cardinale, S., Tueros, F. G. & Sommer, M. O. A. Genetic-metabolic coupling for targeted metabolic engineering. Cell Rep. 20, 1029–1037 (2017).
Shen, B. et al. Fermentative production of Vitamin E tocotrienols in Saccharomyces cerevisiae under cold-shock-triggered temperature control. Nat. Commun. 11, 5155 (2020).
Yuan, P. et al. Engineering a ComA quorum-sensing circuit to dynamically control the production of menaquinone-4 in Bacillus subtilis. Enzyme Microb. Technol. 147, 109782 (2021).
Gao, Q. et al. Highly efficient production of menaquinone-7 from glucose by metabolically engineered Escherichia coli. ACS Synth. Biol. 10, 756–765 (2021).
Zhang, L. et al. Phosphate limitation increases coenzyme Q10 production in industrial Rhodobacter sphaeroides HY01. Synth. Syst. Biotechnol. 4, 212–219 (2019).
Fleige, C., Meyer, F. & Steinbuchel, A. Metabolic engineering of the actinomycete Amycolatopsis sp. strain ATCC 39116 towards enhanced production of natural vanillin. Appl. Environ. Microbiol. 82, 3410–3419 (2016).
Wang, Q. et al. Efficient biosynthesis of vanillin from isoeugenol by recombinant isoeugenol monooxygenase from Pseudomonas nitroreducens Jin1. Appl. Biochem. Biotechnol. 193, 1116–1128 (2021).
Wang, Z., Liu, Z., Cui, W. & Zhou, Z. Establishment of bioprocess for synthesis of nicotinamide by recombinant Escherichia coli expressing high-molecular-mass nitrile hydratase. Appl. Biochem. Biotechnol. 182, 1458–1466 (2017).
Deng, M. D. et al. Metabolic engineering of Escherichia coli for industrial production of glucosamine and N-acetylglucosamine. Metab. Eng. 7, 201–214 (2005).
Lomthong, T. et al. Very high gravity (VHG) bioethanol production using modified simultaneous saccharification and fermentation of raw cassava chips with molasses by Kluyveromyces marxianus DMKU-KS07. Waste Biomass Valoriz. 12, 3683–3693 (2021).
Zhao, Y. et al. High-efficiency production of bisabolene from waste cooking oil by metabolically engineered Yarrowia lipolytica. Microb. Biotechnol. 14, 2497–2513 (2021).
Li, L. et al. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)−2,3-butanediol from lignocellulose-derived sugars. Metab. Eng. 28, 19–27 (2015).
Maina, S. et al. Volumetric oxygen transfer coefficient as fermentation control parameter to manipulate the production of either acetoin or d-2,3-butanediol using bakery waste. Bioresour. Technol. 335, 125155 (2021).
Betterle, N. & Melis, A. Photosynthetic generation of heterologous terpenoids in cyanobacteria. Biotechnol. Bioeng. 116, 2041–2051 (2019).
Gascoyne, J. L., Bommareddy, R. R., Heeb, S. & Malys, N. Engineering Cupriavidus necator H16 for the autotrophic production of (R)-1,3-butanediol. Metab. Eng. 67, 262–276 (2021).
Zheng, Z. Y. et al. An increase of curdlan productivity by integration of carbon/nitrogen sources control and sequencing dual fed-batch fermentors operation. Prikl. Biokhim. Mikrobiol. 50, 44–51 (2014).
Chen, G. et al. Pullulan production from synthetic medium by a new mutant of Aureobasidium pullulans. Prep. Biochem. Biotechnol. 47, 963–969 (2017).
Liu, M. et al. Enhanced bacterial cellulose production by Gluconacetobacter xylinus via expression of Vitreoscilla hemoglobin and oxygen tension regulation. Appl. Microbiol. Biotechnol. 102, 1155–1165 (2018).
Amanullah, A., Satti, S. & Nienow, A. W. Enhancing xanthan fermentations by different modes of glucose feeding. Biotechnol. Prog. 14, 265–269 (1998).
Liu, X. et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 9, 448 (2018).
Niu, W., Kramer, L., Mueller, J., Liu, K. & Guo, J. Metabolic engineering of Escherichia coli for the de novo stereospecific biosynthesis of 1,2-propanediol through lactic acid. Metab. Eng. Commun. 8, e00082 (2019). This study reports an alternative biosynthetic pathway for 1,2-propanediol production that does not generate methylglyoxal, a cytotoxic intermediate of the conventionally used biosynthesis pathway.
Zhang, J. et al. Sesquiterpene synthase engineering and targeted engineering of alpha-santalene overproduction in Escherichia coli. J. Agric. Food Chem. 70, 5377–5385 (2022).
Zhao, S. et al. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab. Eng. 28, 43–53 (2015).
Zhao, X. R., Choi, K. R. & Lee, S. Y. Metabolic engineering of Escherichia coli for secretory production of free haem. Nat. Catal. 1, 720–728 (2018).
Li, Y. W. et al. YALIcloneNHEJ: an efficient modular cloning toolkit for NHEJ integration of multigene pathway and terpenoid production in Yarrowia lipolytica. Front. Bioeng. Biotechnol. 9, 816980 (2021).
Li, Z., Wang, X. & Zhang, H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab. Eng. 54, 1–11 (2019).
Nakano, S., Fukaya, M. & Horinouchi, S. Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti. Appl. Env. Microbiol. 72, 497–505 (2006).
Chen, F., Feng, X., Xu, H., Zhang, D. & Ouyang, P. Propionic acid production in a plant fibrous-bed bioreactor with immobilized Propionibacterium freudenreichii CCTCC M207015. J. Biotechnol. 164, 202–210 (2012).
Hoshino, Y. et al. Stereospecific linalool production utilizing two-phase cultivation system in Pantoea ananatis. J. Biotechnol. 324, 21–27 (2020).
Ignea, C. et al. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate. Nat. Commun. 10, 3799 (2019). This study reports a synthetic biosynthetic pathway involving nerol pyrophosphate to streamline orthogonal production of monoterpenoids without compromising endogenous metabolism using geranyl pyrophosphate.
Connor, M. R., Cann, A. F. & Liao, J. C. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl. Microbiol. Biotechnol. 86, 1155–1164 (2010).
Jang, Y. S. et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio 3, e00314-12 (2012).
Xu, S., Wang, X., Du, G., Zhou, J. & Chen, J. Enhanced production of l-sorbose from d-sorbitol by improving the mRNA abundance of sorbitol dehydrogenase in Gluconobacter oxydans WSH-003. Microb. Cell Fact. 13, 146 (2014).
Saez-Saez, J. et al. Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production. Metab. Eng. 62, 51–61 (2020).
Xu, Y., Zhou, Y., Cao, W. & Liu, H. Improved production of malic acid in Aspergillus niger by abolishing citric acid accumulation and enhancing glycolytic flux. ACS Synth. Biol. 9, 1418–1425 (2020).
Jia, H. et al. Collaborative subcellular compartmentalization to improve GPP utilization and boost sabinene accumulation in Saccharomyces cerevisiae. Biochem. Eng. J. 164, 107768 (2020).
Yang, P. et al. Pathway optimization and key enzyme evolution of N-acetylneuraminate biosynthesis using an in vivo aptazyme-based biosensor. Metab. Eng. 43, 21–28 (2017).
Choi, Y. J., Park, J. H., Kim, T. Y. & Lee, S. Y. Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab. Eng. 14, 477–486 (2012).
Ma, W. et al. Combinatorial pathway enzyme engineering and host engineering overcomes pyruvate overflow and enhances overproduction of N-acetylglucosamine in Bacillus subtilis. Microb. Cell Fact. 18, 1 (2019).
Lee, Y. G. & Seo, J. H. Production of 2,3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae. Biotechnol. Biofuels 12, 204 (2019).
Kim, T. S. et al. Overcoming NADPH product inhibition improves d-sorbitol conversion to l-sorbose. Sci. Rep. 9, 815 (2019).
Jiang, G., Yao, M., Wang, Y., Xiao, W. & Yuan, Y. A “push–pull–restrain” strategy to improve citronellol production in Saccharomyces cerevisiae. Metab. Eng. 66, 51–59 (2021).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Wang, Y., Heermann, R. & Jung, K. CipA and CipB as scaffolds to organize proteins into crystalline inclusions. ACS Synth. Biol. 6, 826–836 (2017).
Eichenberger, M. et al. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab. Eng. 39, 80–89 (2017).
Ignea, C., Pontini, M., Maffei, M. E., Makris, A. M. & Kampranis, S. C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 3, 298–306 (2014).
Peng, B., Plan, M. R., Carpenter, A., Nielsen, L. K. & Vickers, C. E. Coupling gene regulatory patterns to bioprocess conditions to optimize synthetic metabolic modules for improved sesquiterpene production in yeast. Biotechnol. Biofuels 10, 43 (2017).
Gupta, A., Reizman, I. M., Reisch, C. R. & Prather, K. L. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279 (2017).
Wu, Y. et al. Establishing a synergetic carbon utilization mechanism for non-catabolic use of glucose in microbial synthesis of trehalose. Metab. Eng. 39, 1–8 (2017).
Kataoka, N. et al. Enhancement of (R)-1,3-butanediol production by engineered Escherichia coli using a bioreactor system with strict regulation of overall oxygen transfer coefficient and pH. Biosci. Biotechnol. Biochem. 78, 695–700 (2014).
Alonso-Gutierrez, J. et al. Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering. Metab. Eng. 28, 123–133 (2015).
Manjula Rao, Y. & Sureshkumar, G. Direct biosynthesis of ascorbic acid from glucose by Xanthomonas campestris through induced free-radicals. Biotechnol. Lett. 22, 407–411 (2000).
Webb, J. P. et al. Efficient bio-production of citramalate using an engineered Escherichia coli strain. Microbiology 164, 133–141 (2018).
Wu, J., Zhang, X., Zhou, J. & Dong, M. Efficient biosynthesis of (2S)-pinocembrin from d-glucose by integrating engineering central metabolic pathways with a pH-shift control strategy. Bioresour. Technol. 218, 999–1007 (2016).
Gao, M. et al. Efficient endo-beta-1,3-glucanase expression in Pichia pastoris for co-culture with Agrobacterium sp. for direct curdlan oligosaccharide production. Int. J. Biol. Macromol. 182, 1611–1617 (2021).
Inokuma, K., Liao, J. C., Okamoto, M. & Hanai, T. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110, 696–701 (2010).
Li, N. et al. Integrated approach to producing high-purity trehalose from maltose by the yeast Yarrowia lipolytica displaying trehalose synthase (TreS) on the cell surface. J. Agric. Food Chem. 64, 6179–6187 (2016).
Whittaker, J. A., Johnson, R. I., Finnigan, T. J. A., Avery, S. V. & Dyer, P. S. in Grand Challenges in Fungal Biotechnology (ed. Nevalainen, H.) 59–79 (Springer, 2020).
Jang, W. D., Kim, G. B., Kim, Y. & Lee, S. Y. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr. Opin. Biotechnol. 73, 101–107 (2022).
Kim, G. B., Kim, W. J., Kim, H. U. & Lee, S. Y. Machine learning applications in systems metabolic engineering. Curr. Opin. Biotechnol. 64, 1–9 (2020).
Otero-Muras, I. & Carbonell, P. Automated engineering of synthetic metabolic pathways for efficient biomanufacturing. Metab. Eng. 63, 61–80 (2021).
Zhang, J. et al. Accelerating strain engineering in biofuel research via build and test automation of synthetic biology. Curr. Opin. Biotechnol. 67, 88–98 (2021).
Pantoja Angles, A., Valle-Perez, A. U., Hauser, C. & Mahfouz, M. M. Microbial biocontainment systems for clinical, agricultural, and industrial applications. Front. Bioeng. Biotechnol. 10, 830200 (2022).
Arnolds, K. L. et al. Biotechnology for secure biocontainment designs in an emerging bioeconomy. Curr. Opin. Biotechnol. 71, 25–31 (2021).
Gao, C. et al. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat. Commun. 10, 3751 (2019).
Roenneke, B. et al. Production of the compatible solute alpha-d-glucosylglycerol by metabolically engineered Corynebacterium glutamicum. Microb. Cell Fact. 17, 94 (2018).
Bang, H. B., Lee, K., Lee, Y. J. & Jeong, K. J. High-level production of trans-cinnamic acid by fed-batch cultivation of Escherichia coli. Process. Biochem. 68, 30–36 (2018).
Zha, W. et al. Reconstruction of the biosynthetic pathway of santalols under control of the GAL regulatory system in yeast. ACS Synth. Biol. 9, 449–456 (2020).
Wang, C., Park, J. E., Choi, E. S. & Kim, S. W. Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway — application of PgpB and YbjG phosphatases. Biotechnol. J. 11, 1291–1297 (2016).
Hasegawa, S., Tanaka, Y., Suda, M., Jojima, T. & Inui, M. Enhanced glucose consumption and organic acid production by engineered Corynebacterium glutamicum based on analysis of a pfkB1 deletion mutant. Appl. Environ. Microbiol. 83, e02638-16 (2017).
Rhodes, R. A. et al. Production of fumaric acid in 20-liter fermentors. Appl. Microbiol. 10, 9–15 (1962).
Tai, Y. S., Xiong, M. & Zhang, K. Engineered biosynthesis of medium-chain esters in Escherichia coli. Metab. Eng. 27, 20–28 (2015).
Acknowledgements
This work was supported by the Cooperative Research Program for Agriculture Science and Technology Development (project number PJ01577901) from the Rural Development Administration, Republic of Korea, and also by the ‘Development of platform technologies of microbial cell factories for the next-generation biorefineries’ (project number 2022M3J5A1056117) from the National Research Foundation supported by the Korean Ministry of Science and ICT.
Author information
Authors and Affiliations
Contributions
K.R.C. researched data for the article and contributed to the conceptualization, writing, reviewing and editing of the manuscript. S.Y.L. contributed to the conceptualization, reviewing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Sotirios Kampranis, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, K.R., Lee, S.Y. Systems metabolic engineering of microorganisms for food and cosmetics production. Nat Rev Bioeng 1, 832–857 (2023). https://doi.org/10.1038/s44222-023-00076-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-023-00076-y
This article is cited by
-
Use of antioxidants to extend the storage of lyophilized cell-free synthesis system
Biotechnology and Bioprocess Engineering (2024)
-
Biofoundries: Bridging Automation and Biomanufacturing in Synthetic Biology
Biotechnology and Bioprocess Engineering (2023)