Abstract
We propose a deep multi-stream model for left ventricular ejection fraction (LVEF) prediction in 2D echocardiographic (2DE) examinations. We use four standard 2DE views as model input, which are automatically selected from the full 2DE examination. The LVEF prediction model processes eight streams of data (images + optical flow) and consists of convolutional neural networks terminated with transformer layers. The model is made robust to missing, misclassified and duplicate views via pre-training, sampling strategies and parameter sharing. The model is trained and evaluated on an existing clinical dataset (12,648 unique examinations) with varying properties in terms of quality, examining physician, and ultrasound system. We report \(R^2 = 0.84\) and mean absolute error = 4.0% points for the test set. When evaluated on two public benchmarks, the model performs on par or better than all previous attempts on fully automatic LVEF prediction. Code and trained models are available on a public project repository.
Similar content being viewed by others
Introduction
Echocardiography is one of the most common, versatile and cost-effective imaging technique for cardiovascular evaluation1. Estimation of LVEF (left ventricle ejection fraction) is an important part of the assessment of systolic function. It is defined as the percentage of the left ventricle end diastolic volume that is ejected with each contraction and is often calculated with biplane Simpson method2, but also relies on visual assessment “eyeballing” 3. However, evaluation of LVEF with echocardiography is associated with uncertainty because of interobserver variation, with better reproducibility among experienced readers3. Deep learning methods for 2DE analysis can help towards a more automated, consistent and accurate assessment process4.
We propose a deep model for LVEF prediction in 2DE examinations based on an 8-stream convolutional neural network (CNN) and transformer model. We use four 2DE views as input: apical two-, three- and four-chamber (A2C, A3C, A4C), and parasternal long axis (PLAX), which are automatically selected from the full 2DE examination. We focus on 2DE datasets with properties that are common in clinical settings: with varying quality, examining physician and 2DE system, with limited metadata such as missing view information, and with missing or duplicate views. The model is made robust to missing, misclassified and duplicate views via customised pre-training, sampling strategies and parameter sharing. The model is trained, validated and tested on an existing clinical dataset, and in addition, evaluated on two public benchmarks: the EchoNet-Dynamic dataset5, and the CAMUS dataset6. The methods and the results are reported in accordance with the PRIME checklist, see the Supplementary Table S1.7.
There have been several previous attempts to determine LVEF from 2DE examinations with deep models. Some models segment the cardiac chambers in one or several 2DE views, and compute the LVEF from these segmentations6,8,9,10. Others determine LVEF directly from the 2DE examination without an intermediate segmentation step11,12,13,14,15,16, or use a mix of the two approaches5. Most often, LVEF determination is posed as a prediction problem5,6,8,9,10,12,13,14,15,while some pose it as a classification problem11,16. A majority use datasets with known view labels5,6,8,11,13,14,15,16, while only a few address the more challenging problem with unknown views9,10,12. Some only use the A4C view as input5,11,13,15,16, others use both A2C and A4C6,8,9,12, and a few use the five views A2C, A3C, A4C, PLAX and parasternal short axis (PSAX)10,14. Some evaluate their LVEF determination model on publicly available datasets: the EchoNet-Dynamic dataset5,11,15, and the CAMUS dataset6,8,9.
Methods
Model
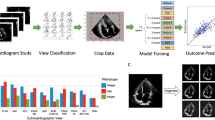
We use four 2DE views as input for the LVEF prediction model: A2C, A3C, A4C and PLAX. These views were selected since they are standard views normally included in most 2DE examinations, and since they contain information that should be helpful for LVEF determination. We use a standard classification model to extract the selected views from the full 2DE examination detailed in the section on pre-processing. For LVEF prediction, we use a 2-stream CNN and transformer model with four views (image + optical flow) as input, that is, eight streams of data in total. See Fig. 1 for a graphical summary of the full model.
The base building block of our LVEF prediction model is the 2-stream I3D model in Carreira and Zissserman, which consists of two 3D CNNs, one for the image and one for the optical flow of the image17. Our reasoning behind using a model originally intended for action recognition is the common denominator of having spatiotemporal data. We modify each 3D CNN by adding a terminating BERT (Bidirectional Encoder Representations from Transformers) layer for temporal pooling, which showed improved results on action recognition tasks in Kalfaoglu et al.18. The motivation for using a terminating BERT layer is to fully exploit the temporal information using transformer attention mechanisms without loosing any information due to averaging and ignored ordering. We use the FRMB (feature reduction by modified block) solution in Kalfaoglu et al. for this18. The same instance of the model (i.e. shared weights) is used to process all the views. We let the four views share parameters since (i) we want the model to be robust to missing, misclassified and duplicate views, and (ii) 2DE views share many common features which allows for a more compact model. The outputs from the eight streams of data are combined in a linear layer. Since each examination might include none, one or several videos of each view class, we construct input data instances according to the following rules: (i) If a view is missing, it is replaced with another view according to the following (descending) priority order: A4C, A2C, A3C, PLAX. This is possible due to the model’s shared weights between views, and should increase the model’s robustness to missing or misclassified views. (ii) If an examination includes several instances of the same view, we create data instances of all possible view combinations. This should increase the model’s robustness to examinations with varying quality, and works as an augmentation strategy. Further, it eliminates the need for a more sophisticated view classifier that chooses between videos of the same view. Details on the optical flow computations can be found in the section on pre-processing.
Data
The study is a retrospective register study. Inclusion criteria were: (i) A 2DE performed at the Department of Clinical Physiology, Sahlgrenska University Hospital, Gothenburg, Sweden between 2007 and 2017, (ii) 2DE performed on a GE ultrasound system (GE Vivid 7, GE Vivid 9, GE Vivid e9), (iii) clinical report signed by an experienced physician with more than 500 signed reports, (iv) saved image data with minimum one of the following views: A2C, A3C, A4C or PLAX, and (v) a numeric value of LVEF in the clinical report. No exclusions were made due to quality issues (reverberations, artefacts, noise). We included all examinations that fulfilled the inclusion criteria and where LVEF was reduced. We balanced the dataset by adding a randomized sample of examinations with normal LVEF. The final dataset consists of 2DE examinations from 12,648 unique patients where LVEF is reduced in 50% of the examinations, supranormal in 1.5%, and normal in 48.5%. Normal LVEF is defined according to Lang et al. as \(\ge 52\%\) for men and \(\ge 54\%\) for women19. Supranormal is defined as \(\ge 70\%\), which is the definition of supranormal used at the site. Since examinations above 70% were classified as 70% (see below), we could not use the definition by Lang et al. (men 72%, women 74%).
Each examination contains 2DE video(s) from one or several views, and corresponding metadata such as heart rate (HR), frames-per-second (FPS), and LVEF. The dataset is split into a training set (70%, 8853 examinations), a validation set (15%, 1898 examinations) and a test set (15%, 1897 examinations). The examinations do not include view metadata, and a view classifier was used to generate view labels for all videos. All examinations include at least one video classified as any of the included views (A2C, A3C, A4C, PLAX). The ground truth LVEF values were manually reported at the time of examination by the examining physician, either by “eyeballing” or by calculations with biplane Simpson or Teichholz method20. The videos differ in size, length and FPS. All LVEF values were at the time of examination reported in multiples of five. LVEF values below 20% were reported as 20%, and LVEF values above 70% were reported as 70%. Note that this procedure of binning and truncation of the LVEF values is the standard practice of the echocardiography lab where the dataset was generated. The study population is summarized in Table 1, and the distribution of LVEF values are reported in Fig. 2. All data was anonymized before use and informed consent was not obtained from the study subjects. This protocol was approved, and the need for informed consent was waived, by the Clinical Medical Research Ethics Board of Sweden (ref. number: 818-18). The study was performed in accordance with this ethical approval and the Declaration of Helsinki. The dataset is described in detail in Hagberg et al.21.
Pre-processing
Videos were converted from RGB to grayscale via averaging, and pixels values were normalized to \([-1, 1]\). Frames were resized to the same size (\(223\times 169\) pixels), and the videos were resampled to have the same FPS/HR, set to 18 frames/heartbeat. This temporal resolution was carefully selected to strike a balance between minimizing computational complexity (favoring lower temporal resolution) and preserving video interpretability (favoring higher temporal resolution). To guarantee that each input video includes one complete cardiac cycle, we used the first 20 frames for each resampled video (corresponding to \(\approx 1.1\) cardiac cycles), and resampled videos with less than 20 frames were periodically extended (“looped”) before resampling. A subset of pre-processed videos was inspected by experienced physicians to make sure that it was possible to analyse LVEF with this pre-processing, and that all inspected videos contained a full heart cycle with both end systolic and end (or near end) diastolic information. We used the Dual TV L1 optical flow algorithm in Zach et al.22. implemented in OpenCV 4.4.0, for flow computations (stopping criterion threshold 0.05, number of warpings per scale 1, number of pyramid scales 1). This is the same optical flow algorithm used by Carreira and Zisserman17, and Kalfaoglu et al.18.
We used a pre-trained Inception2D (v3) from Szegedy et al. for view classification23, where the first convolutional layer was modified to have one input channel (grayscale) instead of three (RGB) by averaging the pre-trained weights. We used five different view classes: (i) A2C, (ii) A3C, (iii) A4C, (iv) PLAX, and (iv) other (which includes all other views). The classes A2C, A3C and A4C include views with optimized depth settings for focusing on the left ventricle. We annotated the view labels for a subset of the examinations in the dataset, and divided these into a training set (70%, 381 examinations), a validation set (10%, 55 examinations) and a test set (20%, 103 examinations). The model was trained on predicting the views from individual frames, the view class for a full video was computed with majority voting. We used the AdamW optimizer24, learning rate (LR) 1e−4, batch size 8, weight decay 2e−5 and the cross-entropy loss (weighted with respect to class distribution). The model reached an average accuracy of 94% for the test set, with class accuracies 95% (A2C), 98% (A3C), 93% (A4C), and 95% (PLAX). Additional view classification networks were also explored, see Hagberg et al., while the best performance was obtained using Inception2D (v3), which is why this architecture was selected21.
Training
We used the training set for learning the weights of the LVEF prediction model, and the validation set for hyperparameter and model selection. The LVEF model was trained with a 3-step procedure: (i) I3D without BERT was pre-trained on ImageNet, we re-used the weights in Kalfaoglu et al.18. (ii) 2-stream I3D with BERT was trained on mixed views, and (iii) 8-stream I3D with BERT was trained on separated views. We used same the training strategies, and the same set of hyperparameters, for step (ii) and (iii). The training data was sampled with respect to continuously updated sample weights computed from the loss for each training data instance. The weight for each training instance was set to the ratio between the training instance loss and the corresponding batch loss, and clamped to [0.1, 3] to avoid extreme sample weights. If an examination in the training set included duplicate views, we sampled one of the possible inputs with an uniform probability over all possible inputs. We did not use any other augmentation strategies since evaluated techniques (e.g. using different types of noise, geometric transformations, brightness adjustments, occlusion strategies) gave no significant improvements on the validation set. We used the AdamW optimizer24, LR 1e−3, batch size 16, weight decay 1e−4, loss function MSE, half-precision accuracy for input data and a LR scheduler that decreases the LR by a factor 1e1 if no improvement on the validation loss can be seen after 100 iterations (validation loss were computed every 10th iteration). If an examination in the validation set included duplicate views, we used weighting when computing the evaluation metrics such that each examination had a total weight equal to one. For all experiments, we used PyTorch 1.7.1 with Cuda 10.1. Average training time was 136 h (500 iterations) on a Nvidia DGX-2 with 22 CPUs and four V100 GPUs, with a memory footprint of 218 GB RAM and 19 GB per GPU.
Evaluation
We evaluated the model’s performance on an internal test set with 1897 examinations, on two public benchmark datasets and in an ablation study investigating the impact of (i) sharing the weights between the models processing each view, (ii) replacing missing views with other view classes according to the predefined priority order, (iii) using all available view instances in case of duplicate views, (iv) using optical flow as a second data stream, (v) using BERT as a terminating layer, (vi) different loss functions and (vii) different number of input views. We used bias ± standard deviation (SD), the coefficient of determination \(R^2\) and the mean absolute error (MAE) as evaluation metrics.
Results
Test set results
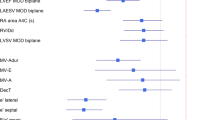
We used the 1897 examinations in the test set for testing the model’s final performance. If an examination in the test set included duplicate views, we used averaging over all possible outputs to compute the LVEF prediction. See Table 2 for bias ± SD, \(R^2\) and MAE depending on the available input views, and Fig. 3 for scatter and Bland-Altman plots.
CAMUS and EchoNet-dynamic results
Table 3 reports the model’s performance when evaluated on the CAMUS and the EchoNet-Dynamic test datasets, as well as previously reported LVEF prediction results for these datasets.
For the CAMUS dataset, we finetuned and evaluated a 4-stream version of the model, since the CAMUS dataset only includes A2C and A4C views. We used the training dataset for finetuning (450 examinations) and the test dataset for testing (50 examinations). We employed the model trained on our in-house dataset for initialization, and used the identical learning strategy and hyperparameters as outlined in the Training section. The CAMUS examinations only include the contraction phase (systole), and lack heart rate metadata. Therefore, we resampled all videos to have 20 frames corresponding to one contraction.
For the EchoNet-Dynamic dataset, we finetuned and evaluated a 2-stream version of the model, since the dataset only includes A4C views. We used the training dataset for finetuning (10751 examinations) and the test dataset for evaluation (1897 examinations). Since the EchoNet-Dynamic examinations lack heart rate metadata, we resampled all videos to have 20 frames with 9 frames/second. As for the CAMUS dataset, we used the model trained on our in-house dataset for initialization, and the same learning strategy and hyperparameters as outlined in the Training section.
Ablation study
Table 4 reports \(R^2\) and MAE values for the following comparisons on the validation set: (i) w/o shared model weights between the different views, (ii) w/o view replacement in case of missing views by using empty views instead, (iii) w/o using all available views in case of duplicate views, (iv) w/o optical flow as a second data stream in the I3D models, (v) w/o BERT as a terminating layer in the I3D models, (vi) with MSE (regression), balanced all-threshold (ordinal regression) and balanced cross-entropy (classification) as loss function, and (vii) with one input view (A4C) in a 2-stream model, two input views (A2C, A4C) in a 4-stream model, three input views (A2C, A3C, A4C) in a 6-stream model, and four input views (A2C, A3C, A4C, PLAX) in an 8-stream model. To ensure a fair comparison, the LVEF predictions are rounded to the nearest multiple of five for the MSE loss in comparison (vi), and the validation set only includes examinations with at least one A4C (1870 examinations) in comparison (vii).
Discussion
In this paper, we present a fully automatic model for LVEF assessment, with a MAE of 4.0 pp. We conclude from Table 2 that while depending on all included views for optimal performance, the model relies less on A2C, A3C and A4C than on PLAX, which is expected due the somewhat overlapping information (from the same apical window) in A2C, A3C, and A4C compared to PLAX using a different ultrasound window. One possible reason for the less impressive performance for LVEF = 20% and 70% than the intermediate values in Fig. 3 is the truncation of the ground truth labels, which may have resulted in a model prone to overestimate small LVEF values, and correspondingly, to underestimate large LVEF values.
Second, we conclude that using BERT as a terminating layer is crucial for optimal performance, which is expected due to transformers’ superior capability to incorporate temporal information compared to e.g. average or max pooling. Further, we conclude that using all possible view instances in case of duplicate views boosts the model’s performance, which is of little surprise since it increases the amount of available training data. Further, the model’s performance consistently improves with an increased number of views, and using optical flow seems to boost the performance somewhat. However, these improvements (more views, and multiple data streams) need to be weighted against the increased computational complexity they add. Using view replacement gives a modest performance boost, however, it has the advantage of adding no computational complexity. Similarly, using shared weights between the different views only gives a small performance boost, while it has the advantage of significantly reducing the computational complexity. Surprisingly, there is only small differences between posing LVEF determination as a regression, ordinal regression and classification problem.
When comparing the proposed model to previous methods on the CAMUS dataset, we can conclude that we outperform all previous works in terms of bias, MAE, \(R^2\) and SD. When comparing the proposed model to previous methods on the EchoNet-Dynamic dataset, we can conclude that we outperform one, and perform on par with another, in terms of MAE and \(R^2\). Our dataset comprises real-world clinical examinations from hospital archives, with no exclusions based on image quality. We view this as a strength, as more curated datasets, such as CAMUS, tend to present a simplified problem. Conducting a prospective test in a clinical setting would undoubtedly provide a valuable opportunity to further assess performance.
We envision (at least) three immediate research directions. Firstly, when evaluating, the video most suitable for LVEF determination should be automatically selected when there are duplicate views, instead of using weighting/averaging. Secondly, the output from each stream should be paired with a learned model confidence to enable a more sophisticated fusion of the output from each stream. Finally, the model should be implemented in a picture archiving and communication system, and evaluated in a clinical setting, including analysis of the clinical significance, usability and reliability.
Limitations
Our paper has several limitations. (i) One limitation arises from the nature of our dataset, a real-world clinical dataset. Specifically, LVEF values in this dataset have been truncated, with values below 20% set to 20% and values exceeding 70% set to 70%. While this truncation aligns with clinical practices, it introduces a potential drawback in our study. It may lead to an overestimation of low LVEF values and an underestimation of high LVEF values. It’s important to acknowledge, however, that the clinical relevance of this limitation is likely minimal. (ii) Another limitation stems from the fact that we did not explore various methods for balancing the dataset in favor of impaired LVEF values. Employing such balancing techniques could have potentially improved our results, particularly for low LVEF values. (iii) A further limitation pertains to the handling of duplicate views during examinations. In our analysis, we incorporated all possible inputs from duplicate views and averaged the LVEF measurements obtained from them. This approach deviates from a more refined dataset, where only the best-case duplicate view is selected. However, it is essential to note that our analysis is fully automated, and incorporating a manual step to choose the optimal duplicate view is not a practical option. In summary, while our study is not devoid of limitations, we believe that these constraints do not significantly detract from the clinical implications of our findings.
Conclusions
We have proposed a deep 8-stream model for LVEF prediction in 2DE examinations using four automatically selected 2DE views as input. The model was trained and evaluated on an existing clinical dataset with varying quality, examining physician and 2DE system, with limited metadata such as missing view information, and with missing or duplicate views. We reported \(R^2 = 0.84\), MAE = 4.0% points and bias = 0.1 ± 5.2% points for the test set. We also evaluated on two public benchmarks. These datasets differ significantly from our focus: they include only A2C/A4C and A4C views respectively, no examinations have missing or duplicate views, and view labels are known. Still, we performed on par or better than all previous LVEF prediction methods evaluated on these two datasets.
Data availability
The in-house dataset analysed during the current study are not publicly available since the ethical approval does not allow for this. However, the dataset is available from the corresponding author on reasonable request including an approved ethical approval by the Clinical Medical Research Ethics Board of Sweden. The two benchmark datasets are publicly available at the CAMUS project homepage and EchoNet Dynamic homepage. The trained models are available on a public project repository.
References
Papolos, A., Narula, J., Bavishi, C., Chaudhry, F. A. & Sengupta, P. P. US hospital use of echocardiography: Insights from the nationwide inpatient sample. J. Am. Coll. Cardiol. 67, 502–511 (2016).
Schiller, N. B. et al. Left ventricular volume from paired biplane two-dimensional echocardiography. Circulation 60, 547–555 (1979).
Kouris, N. T. et al. Left ventricular ejection fraction and global longitudinal strain variability between methodology and experience. Echocardiography 38, 582–589 (2021).
Litjens, G. et al. State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc. Imaging 12, 1549–1565 (2019).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020).
Leclerc, S. et al. Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans. Med. Imaging 38, 2198–2210 (2019).
Sengupta, P. P. et al. Proposed requirements for cardiovascular imaging-related machine learning evaluation (PRIME): a checklist— reviewed by the American College of Cardiology Healthcare Innovation Council. Cardiovasc. Imaging 13, 2017–2035 (2020).
Liu, X. et al. Deep learning-based automated left ventricular ejection fraction assessment using 2-D echocardiography. Am. J. Physiol. Heart Circ. Physiol. 321, H390–H399 (2021).
Smistad, E. et al. Real-time automatic ejection fraction and foreshortening detection using deep learning. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 67, 2595–2604 (2020).
Zhang, J. et al. Fully automated echocardiogram interpretation in clinical practice: Feasibility and diagnostic accuracy. Circulation 138, 1623–1635 (2018).
Esfeh, M. M. K., Luong, C., Behnami, D., Tsang, T. & Abolmaesumi, P. A deep bayesian video analysis framework: Towards a more robust estimation of ejection fraction. in International Conference on Medical Image Computing and Computer-Assisted Intervention 582–590 (Springer, 2020).
Behnami, D. et al. Dual-view joint estimation of left ventricular ejection fraction with uncertainty modelling in echocardiograms. in Medical Image Computing and Computer Assisted Intervention696–704 (Springer, 2019).
Ghorbani, A. et al. Deep learning interpretation of echocardiograms. NPJ Digit. Med. 3, 1–10 (2020).
Kusunose, K. et al. Deep learning for assessment of left ventricular ejection fraction from echocardiographic images (J. Am. Soc, Echocardiogr, 2020).
Reynaud, H. et al. Ultrasound video transformers for cardiac ejection fraction estimation. in Medical Image Computing and Computer Assisted Intervention 495–505 (Springer, 2021).
Silva, J. F., Silva, J. M., Guerra, A., Matos, S. & Costa, C. Ejection fraction classification in transthoracic echocardiography using a deep learning approach. in 2018 IEEE 31st International Symposium on Computer-Based Medical Systems 123–128 (IEEE, 2018).
Carreira, J. & Zisserman, A. Quo vadis, action recognition? A new model and the kinetics dataset. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 6299–6308 (2017).
Kalfaoglu, M. E., Kalkan, S. & Alatan, A. A. Late temporal modeling in 3D CNN architectures with BERT for action recognition. in Computer Vision ECCV 731–747 (Springer, 2020).
Lang, R. M. et al. Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur. Heart J. Cardiovasc. Imaging 16, 233–271 (2015).
Teichholz, L. E., Kreulen, T., Herman, M. V. & Gorlin, R. Problems in echocardiographic volume determinations: Echocardiographic–angiographic correlations in the presence or absence of asynergy. Am. J. Cardiol. 37, 7–11 (1976).
Hagberg, E. et al. Semi-supervised learning with natural language processing for right ventricle classification in echocardiography-a scalable approach. Comput. Biol. Med. 143, 105282 (2022).
Zach, C., Pock, T. & Bischof, H. A duality based approach for realtime TV-L 1 optical flow. in Joint Pattern Recognition Symposium 214–223 (Springer, 2007).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2818–2826 (2016).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101 (2017).
Acknowledgements
We wish to thank Nasser Hosseini and Anders Arvidsson for data preparation. The study was financed by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement, as well as grants from Analytic Imaging Diagnostics Arena (AIDA), Sahlgrenska University Hospital, Gothenburg Medical society and Chalmers University of Technology.
Funding
Open access funding provided by Chalmers University of Technology.
Author information
Authors and Affiliations
Contributions
J.A. and O.H. proposed the main idea. D.H. and E.H. proposed several method components related to the implementation and the data, respectively. J.A., D.H. and R.P. implemented the method and conducted the experiments. E.H and O.H. curated and annotated the data. J.A., O.H., E.H. and D.H. analyzed the results. J.A. wrote the main parts of paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alvén, J., Hagberg, E., Hagerman, D. et al. A deep multi-stream model for robust prediction of left ventricular ejection fraction in 2D echocardiography. Sci Rep 14, 2104 (2024). https://doi.org/10.1038/s41598-024-52480-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52480-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.