Abstract
The Vietnamese indica landrace ‘Tetep’ is known worldwide for its durable and broad spectrum-resistance to blast. We performed genetic and molecular analyses of leaf blast resistance in a Tetep derived recombinant inbred line ‘RIL4’ which is resistant to both leaf and neck blast. Phenotypic analysis of segregating F2 progenies suggested that leaf blast resistance in RIL4 was controlled by a dominant gene tentatively designated as Pi-l(t). The gene was mapped to a 2.4 cm region close to the centromere of chromosome 12. The search for the gene content in the equivalent genomic region of reference cv. Nipponbare revealed the presence of five NBS-LRR genes, two of which corresponded to the alleles of Pita and Pi67 genes previously identified from Tetep. The two other genes, LOC_Os12g17090, and LOC_Os12g17490 represented the homologs of stripe rust resistance gene Yr10. The allelic tests with Pita2 and Pi67 lines suggested that the leaf blast resistance gene in RIL4 is either allelic or tightly linked to these genes. The genomic position of the leaf blast resistance gene in RIL4 perfectly coincided with the genomic position of a neck blast resistance gene Pb2 previously identified from this line suggesting that the same gene confers resistance to leaf and neck blast. The present results were discussed in juxtaposition with past studies on the genes of Pita/Pita2 resistance gene complex.
Similar content being viewed by others
Introduction
Rice [Oryza sativa (L.) 2n = 2x = 24] belongs to genus Oryza and family Poaceae. The genus is composed of 24 species, out of which two species O. sativa L. and O. glaberrima are cultivated1. Its high energy digestibility coupled with fact that it serves as the primary calorie source for nearly the one third of world’s population makes rice one of the most important cereal crops in the world2,3. During 2021 rice has been cultivated in an area of 165.25 million hectares in the world with annual production of 787.29 million tonnes4. With the current rate of population growth, the global rice demand is expected to escalate to 8.52 × 108 tonnes by 2035 requiring further genetic improvements5. The yield gains achieved through plant breeding are frequently offset by the damage inflicted by various biotic and abiotic stresses allowing farmers to realize only half of the rice yield potential6. Among the biotic stresses, blast disease caused by hemibiotrophic ascomycete fungus Pyricularia oryzae Cavara (Telomorph, Magnaporthe oryzae), is one of the most widespread and destructive diseases. The disease results in yield loss of $66 billion worldwide annually, which is enough to feed 60 million people7. The disease affects crop at almost all growth stages leading to two easily recognizable phases of the disease: leaf blast occurring during the vegetative stage causing spindle shaped lesions on leaf blades and leaf collars, and neck blast (synonymous with panicle blast) which is characterized by infection at plant nodes and different parts of panicle and grains. The neck blast is economically more significant because a single infection at the panicle base can result in completely empty panicles, and losses up to 70% have been recorded in the crop affected by neck blast8. Despite the availability of many potent fungicides, potential environmental and health risks along with economic considerations make chemical control of the disease an ungainly prospect. Under such constraints, utilization of host resistance mediated by resistance genes is the most efficient practice to manage the disease. Identification of genes for resistance to leaf and neck blast and their utilization in breeding resistant varieties is the most efficient management practice to manage the disease.
Until now some 120 blast resistance genes have been identified and 25 of those have been cloned and molecularly characterized9,10,11,12,13,14. While a great many blast resistance genes identified till date are effective against leaf blast, only three, namely, Pb1, Pi25 and Pi64 are known to confer resistance to neck blast phase of the disease15,16,17. Reported susceptibility to one phase of disease in genotypes resistant to the other phase indicates that the mechanisms and underlying genetic factors involved in neck and leaf blast resistance may be different8,18,19. Under such circumstances the identification of genes that confer simultaneous resistance to both leaf and neck blast phases of the disease is urgently required for effective management of the disease.
The Vietnamese indica landrace ‘Tetep’ has shown remarkably broad spectrum and persistent resistance against various strains of the blast pathogen in different parts of the world20,21. The genotype has shown enduring resistance to both leaf and neck blast disease for several years in north-western Indian states of Himachal Pradesh, Uttarakhand and J&K22. Tetep has been widely adopted as the progenitor variety in several breeding programs due to its durable and broad spectrum resistance profile23. Till date six leaf blast resistance genes viz., Pi-1, Pi-kh and Pi54 on chromosome 11, and two putatively allelic genes Pita and Pita2 and Pi67 on chromosome 12 have been identified from the Tetep24,25,26,27,28,29. A new neck blast resistance gene, Pb2 mapped to centromeric region of chromosome 12 has recently been identified from a recombinant inbred line RIL4 that derives its resistance from Tetep30. These studies suggested that the genomic region close to centromere of chromosome 12 of Tetep harbors a cluster of resistance genes that can exploited in rice breeding for developing cultivars resistant to both leaf and neck blast phase of the disease. The efficient exploitation of this region in resistance breeding, however, would require precise dissection of resistance spectrum of these genes to identify the genes that provide resistance to both leaf and neck blast phases the disease. In present study, we performed the genetic and molecular analyses of leaf blast resistance of RIL4, a recombinant inbred line derived from Tetep, that has previously been shown to harbor a neck blast resistance gene Pb2 in order to clarify whether the same gene confers resistance to both leaf and neck blast or there are different genes for resistance to different phases of the disease. The allelic relationship of leaf blast resistance gene identified from RIL4 to leaf blast resistance genes Pita2 and Pi67 previously identified from the same region of Tetep was also investigated. The genetic mapping of leaf blast resistance gene from RIL4 will ensure effective and precise manipulation leaf and neck blast resistance gene(s) identified from this genotype in breeding blast resistant varieties.
Results
Genetic analysis of leaf blast resistance
Altogether nine hundred and thirty-four F2 plants derived from a cross between HPU2216 and blast resistant genotype RIL4 were inoculated with blast isolate Po-HPU2216-5-2. The genotype HPU2216 with Pita gene displayed susceptibility to leaf blast, while RIL4 was completely resistant showing no symptoms of the disease (reaction type 0) (Fig. 1). Of the total inoculated F2 seedlings, 683 exhibited resistant reaction, while 251 were susceptible to leaf blast. The segregation of resistant and susceptible plants showed a good fit to segregation ratio expected for a single dominant gene (Table 1). The new leaf blast resistance gene identified from RIL4 was tentatively designated as Pi-l(t).
Mapping of leaf blast resistance gene
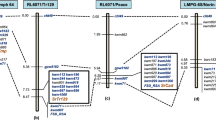
Since a single dominant gene Pb2 located on chromosome 12 has previously been shown to confer neck blast resistance in line RIL430, we initiated mapping of leaf blast resistance gene using the polymorphic markers from the same chromosome on premise that the leaf blast resistance may be conditioned by the similar gene. Initially, two polymorphic markers STS-5 and RM1261, one each from the short and long arm of chromosome 12, were tested for their linkage to leaf blast resistance gene in a mapping population comprising of 220 susceptible F2 plants of cross HPU2216 x RIL4. A total of 13 recombination events were detected between STS-5 and resistance gene, whereas 7 single cross events were detected with RM1261 in a population of 220 susceptible F2 plants (equivalent to 440 F1 gametes) thereby suggesting that the leaf blast resistance gene is also located on chromosome 12 (Supplementary Table S1; Supplementary Fig. S1). Since the recombinants detected with STS-5 and RM1261 were non-identical, these markers were inferred to flank the gene at a distance of 2.9 and 1.5 cm, respectively. Based on this data, the resistance gene was initially mapped to a 4.4 cm genetic interval flanked by STS-5 and RM1261 on chromosome 12 (Fig. 2). To further narrow down the genomic region for the resistance gene, chromosome walking was initiated with the markers located within the interval STS5-RM1261. Of the 11 recombinants detected with distal marker STS-5, 9 were detected by RM3246 marker, whereas of the 7 recombination events detected with RM1261, only three were detected at RRS69 locus (Fig. 2). Of the three recombinants detected with RRS 69, two were also detected with RRS19 (Fig. 2). With this analysis the genomic region for leaf blast resistance gene Pi-l(t) was eventually narrowed down to a 2.4 cm region defined by RM3246 located at 2.0 cm on telomeric side and RRS19 located at 0.4 cm from the gene on centromeric side. Within this narrow genetic interval three SSR markers viz., RRS12, RRS77 and RRS16 co-segregated with the resistance gene as no recombination could be detected between these markers and Pi-l(t) (Supplementary Fig S2).
Genetic and physical map of leaf blast resistance gene identified from RIL4. (a) The genetic map of Pi-l(t) gene based on linkage analysis of F2 progeny of cross HPU2216 x RIL4. *The numbers of recombination events obtained between the relevant markers and Pi-l(t). The numbers to the left of map are relative genetic distances in centimorgans. (b) The physical map of Pi-l(t) gene constructed by e-landing of closely linked markers on the genome sequence of cv. Nipponbare (IRGSP 1.0) released by International Rice Genome Sequencing Project (http://rapdb.dna.affrc.go.jp). The dotted lines designate the positions of the markers on the genome of cv. Nipponbare. The numbers to the right of the map are the distances between the markers in Mbp. The physical positions of Pita3(t) (9.88–10.06), Pita (10.60), Pi67(t) (10.61–12.09), Pi-42(t) (8.07–12.24) and Ptr (10.71–10.98) in million base pairs (Mb) were are adapted from Chen et al.31, Jia and Martin32, Joshi et al.29, Kumar e al.33 and Meng et al.34. (c) Candidate resistance genes with NBS-LRR features identified in the region of Pi-l(t) by searching the TIGR Rice Genome Annotation Project database of cv. Nipponbare (http://rice.plantbiology.msu.edu). CEN: Centromere.
Physical mapping and identification of resistance gene candidates
Based on chromosome landing of the flanking markers RM3246 and RRS19 on the genome of reference cultivar Nipponbare, the leaf blast resistance gene has been delimited to 6.19 Mb region spanning from position 9,095,272 to 15,287,816 bp close to centromere of chromosome 12 (Fig. 2). The search for gene content in the equivalent genomic region of reference cv. Nipponbare (http://rice.plantbiology.msu.edu) revealed the presence of 334 putative genes, of which five namely, LOC_Os12g17090, LOC_Os12g17490, LOC_Os12g18360, LOC_Os12g18374 and LOC_Os12g25170 had features of typical disease resistance genes encoding Nucleotide-binding site and leucine-rich repeat (NB-LRR) proteins (Table 2, Fig. 2). Two of these genes namely, LOC_Os12g17090, and LOC_Os12g17490 represent the homologs of stripe rust resistance gene Yr10, while LOC_Os12g18360 corresponds to a well known blast resistance Pi-ta gene that has been previously been reported to be present in Tetep, the progenitor of donor line RIL-4 . The locus LOC_Os12g18374 corresponds to an allele of blast resistance gene Pi-67(t) gene that has previously been identified from Tetep29. All the five candidate resistance genes identified in the Pi-l(t) region are functional in rice genome as the expression evidences in the form of FL-cDNA transcripts and proteins have been detected for all the genes in the Rice Annotation Project database (http://rapd.dna.affrc.go.jp) (Table 2). The genomic position of the leaf blast resistance gene perfectly coincided with the genomic position of the neck blast resistance gene Pb2 previously identified from the RIL4. Both the genes have been mapped to a 6.19 Mb region flanked by RM3246 and RRS19 and perfectly co-segregate with SSR markers RRS12, RRS16 and RRS17. These results strongly suggest that the same gene confers resistance to leaf and neck blast in RIL4.
Allelic relationship of resistance gene with Pita 2 and Pi67
Previously two leaf blast resistance genes Pita/Pita2 (presumed to be as allelic or tightly linked) and Pi67 have identified from the same genomic region of chromosome 12 of Tetep29,35. The Locus, LOC_Os12g18374 which is one of the five candidates for leaf blast resistance gene identified from RIL4, has also been shortlisted as a candidate for leaf blast resistance gene Pi-67 identified from TDH25129. Therefore, to see whether the leaf blast resistance gene identified from RIL4 is identical to Pita/Pita2 and Pi-67 genes, its allelic relationship with these genes was studied by observing the segregation of leaf blast resistance in F2 progenies of crosses of RIL4 with IRBLta2-Pi and TDH251, respectively. No susceptible individual was found among the F2 progenies of crosses RIL4 x TDH251 and RIL4 x IRBLta2-Pi thereby suggesting that the leaf blast resistance gene in RIL4 is either identical or tightly linked to Pita/Pita2 or Pi67 (Table 3).
Discussion
The blast disease affects rice crop from seedling to reproductive stage thus necessitating the identification of all-stage effective resistance genes that could provide protection throughout the crop season. A great majority of blast resistance genes identified till date have been identified through phenotypic analysis of progenies of different crosses for leaf blast resistance. Owing to inherent difficulties associated with phenotyping of neck/panicle blast resistance, a few major genes mediating protection to neck blast have been identified; of some 118 major blast resistance genes identified from rice, only three, Pb1, Pi25 and Pi64, are known to provide resistance to panicle and neck blast15,16,17. There has been a difference of opinion among the rice researchers regarding the relationship between neck and leaf blast resistance genes. While some researchers have argued that the mechanisms of leaf and neck blast resistance are different based on the fact that genotypes showing resistance to leaf blast in some instances are susceptible to neck blast and vice versa18,19,36. Others have inferred that same genes are involved in providing resistance against both phases of the disease17,37.
Similar to our results, two more blast resistance genes Pi25(t) and Pi64 have previously been shown to confer resistance to both leaf and neck blast, and in many instances the gene/QTLs for panicle blast resistance have co-localized with major leaf blast resistance genes37, thereby suggesting the involvement of common genes for resistance to both leaf and neck blast.
The leaf blast resistance gene identified in RIL4 has been mapped to a recombination suppressed region near the centromere of chromosome 12 from where three blast resistance genes, namely, Pita, Pita2 and Pi67 have previously been identified in Tetep, the resistance donor for line RIL429,35,38. The fact that the rice variety HPU2216, used as a susceptible parent in our study, contains Pita gene inherited from a popular rice variety IR36 suggests that this gene is not involved in blast resistance under our conditions. On the other hand, the lack of segregation of resistance in F2 progenies of crosses of RIL4 with IRBLta2-Pi and TDH251 has suggested that the leaf blast resistance gene in RIL4 is either allelic or tightly linked to Pita2 and Pi67 genes previously identified from the same region29,35.
The genomic region close to centromere of chromosome 12 is known to harbor a cluster of blast resistance genes and nearly 19 allelic and/or closely linked blast resistance genes have been mapped to this region in different rice genotypes39,40. The same genomic region in Tetep and other broad-spectrum resistance genotype Tadukan has been shown to harbor three blast resistance genes Pita, Pita2, Pi6729,35,38. The gene Pita was the first blast resistance genes to be identified from the region followed by Pita224,25. Subsequent genetic studies have indicated that Pita and Pita2 are allelic or closely linked38. Literature reports have consistently suggested that the Pita2 exhibits a broader spectrum of resistance compared to Pita, the former controlling all the strains controlled by Pita plus additional strains that are virulent to Pita; till date no isolate that is virulent on Pita2 and avirulent to Pita has been recorded25,35,38. These observations have been taken to reflect that the Pita2 specificity is a combination of at least two R genes- Pita plus a second linked R-gene38,41. All the Pita2 rice varieties analyzed till date like Tetep, Reiho, Katy and isogenic lines namely, IRBLta2-Re and IRBLta2-Pi are reported to contain resistant Pita allele suggesting that Pita is required for resistance function of Pita238,41. Another gene Ptr located in the linkage block near the centromere of Chromosome 12 has been identified in the U.S. tropical japonica cultivar Katy harboring Pita/Pita2genes inherited from the Tetep. The mutants with defects in Ptr gene are compromised in Pita and Pita2 resistance thereby indicating that the gene is involved in transduction of defense signaling in Pita/Pita2-mediated resistance pathway32. The Ptr gene has been cloned recently and shown to encode a protein with an Armadillo (ARM) repeat domain41. Interestingly, all the rice varieties identified to display Pita2 resistance till date invariably harbor a Katy type resistant Ptr allele in addition to having resistant Pita allele41. The rice variety Yashiro-mochi in which Pita gene was initially cloned38 and few other Pita rice varieties like Pi No.1, Pi No.2 and YT14 incidentally do not harbor resistant Ptr haplotype similar to Pita2 rice varieties41. These data suggest that Pita2 resistance specificity is conditioned by a combination of Pita, Ptr and yet unknown R-gene that probably recognizes the isolates virulent on Pita gene, while the Pita mediated resistance in varieties like Yashiro-mochi operates through a different pathway that does not require Ptr protein as a component of defense signaling pathway.
The largest class of R genes in plants belongs to the conserved family of nucleotide-binding site leucine–rich repeat (NBS-LRR) genes that play role in detecting pathogen effectors (avirulence proteins) and activate defense signaling. Approximately 500 NBS-LRR genes have been predicted in the rice genome, and all the major blast resistance genes cloned until date, except for Pid2, belong to this class39. Although the majority of these genes were identified as single loci that follow Flor’s gene-for-gene model, the blast resistance in several cases has been shown to be mediated by functional pairs of NBS-LRR genes as seen for resistances mediated by Pia, Pi5 and Pik42,43,44. Such NBS-LRR gene pairs show extremely tight physical linkage and are arranged in inverted orientation suggesting a common mechanism of their action45. In such gene pairs, one member known as “sensor” functions to negatively regulate the other member known as “helper” in an inactive complex to prevent autoimmunity in the absence of pathogen, and binding of the pathogen AVR gene encoded effectors triggers the release of this negative regulation allowing the helper to activate downstream defense signaling46. Autoactive NBS-LRR helper and their negative regulators are expected to function as a single unit and likely to remain genetically linked47. Further, emerging evidences in different crops suggest that NBS-LRR genes form genetic networks in which a single helper gene can interact with different sensors that confer immunity to diverse pathogens e.g. a single helper NRC4 is required for the function of several sensors NBS-LRR genes that confer immunity to oomyctes, bacteria, viruses nematodes and insects47.
Genome wide analysis of NBS-LRR genes in Tetep has also revealed the presence of 43 NBS-LRR gene pairs comprising of closely linked members that are arranged in head-to-head orientation in the genome21. One such pair of NBS-LRR genes represented by functional genes LOC_Os12g18360 and LOC_Os12g18374 is also located in the Tetep derived region associated with leaf and neck blast resistance in RIL4. The gene locus LOC_Os12g18360 corresponds to the blast resistance gene Pita, while LOC_Os12g18374 is reported to be a candidate for two different leaf blast resistance genes, Pi67 in Tetep29 and Pi42 in rice genotype DHR933. The CRISPR/Cas9 knockouts of LOC_Os12g18360 (Pita) in transgenic lines expressing matching partner LOC_Os12g18374 are also reported to exhibit auto-immunity thus suggesting that the two genes work as a sensor-helper pair21.
Taking cue from the findings of earlier studies on Pita2 gene25,35,38,41 and current developments in understanding of disease resistance in plants21,46,47, we propose that Pita2 resistance specificity is conditioned by at least three tightly linked NBS-LRR genes. Among these, the helper encoded by LOC_Os12g18374 interacts with two different sensors, one encoded by gene LOC_Os12g18360 (Pita) which retains recognition specificity for effectors of Pita avirulent isolates, and other encoded by one of three closely linked NBS-LRR genes located in the Pi-l(t) region which recognizes the effectors of Pita virulent isolates. Further studies involving generation of gene knockouts of candidate NBS-LRR genes in RIL4 and testing their resistance spectrum against a diverse collection of Pita and Pita2 specific blast isolates are required to explain the molecular basis of Pita2 resistance specificity. The mapping of leaf blast resistance gene in RIL4 to the same Tetep derived genomic region as was previously shown to be associated with neck blast resistance suggested that the same gene (Pi-l(t) = Pb2 = Pita2), confers resistance to both phases of disease. The gene is located in a recombination suppressed region harboring a cluster of NBS-LRR genes that all are functionally active in rice genome as evinced by FL-cDNA and protein expression support for these genes in Nipponbare genome. The gene along with its linked NBS-LRR genes can be effectively incorporated into susceptible genetic backgrounds using co-segregating markers RRS12, RRS77 and RRS16 to develop leaf and neck blast resistant varieties.
Methods
Plant material and resistance phenotyping
An indica rice variety, HPU2216, containing blast resistance gene Pita was used as a susceptible parent. The variety is derived from the cross IR8/IR 2053–521-1–1//IR36 and inherit its Pita gene from the popular IRRI bred rice variety IR36. RIL4, an F2:12 recombinant inbred derivative of cross HPU2216 x Tetep from which a neck blast resistance gene Pb2 has been mapped previously30 was used as a resistant parent. An F2 population derived from the cross of HPU2216 with RIL4 was used for genetic studies and mapping of leaf blast resistance gene. The seeds of parental genotypes and F2 population were sown in a plastic trays (40 × 25 × 10 cm) filled with potting mixture (soil: sand: 3: 1) and grown in a growth chamber maintained at 25 to 30 °C for 21 days.
The P. oryzae isolate, Po-HPU2216-5–2 (race U63-i0-k167-z04-ta023), which is virulent on HPU2216 and avirulent on RIL4 and Tetep was used for the phenotypic screening of leaf blast resistance. The virulence spectrum of the isolate on monogenic blast differentials is provided in Supplementary Table S2. Altogether 934 F2 seedlings of cross HPU2216 x RIL4 were spray inoculated with the culture of this isolate. The procedure described by Rathour et al.48 was used for the preparation of inoculum and disease scoring. Briefly, mycelia from 10-day old cultures were macerated in 5 ml of distilled water and plated onto Mathur’s medium49 for sporulation in Petri plates. After 8 to 10 days of incubation at 25 ± 1 °C, the plates were washed with 10 ml of distilled water to make spore suspension, which was filtered through two layers of muslin cloth and spore concentration adjusted to 40–50 conidia/microscopic field (40 X). About 30–40 ml of the spore suspension was sprayed on 21-day-old seedlings using a glass atomizer. The inoculated seedlings were kept in a humidity chamber at 25 ± 1 °C and sprayed three to four times a day with distilled water to maintain high humidity. Disease reaction was recorded after 7 days of inoculation using 0–5 scale given by Mackill and Bonman26. The number of resistant and susceptible F2 seedlings was recorded and the data subjected to Chi-square analysis to test the goodness of fit to Mendelian ratios. The individual seedlings showing highly resistant (0–1 score) or susceptible (4–5 score) reactions were transferred to pots for later use in extraction of DNA. The DNA from 220 susceptible F2 plants was used for the mapping of the resistance gene using recessive class analysis50.
Mapping of leaf blast resistance gene
Genomic DNA of the parents and F2 seedlings was isolated using the standard CTAB method51. Initially, two polymorphic markers STS-5 and RM1261, one each from the short and long arm of chromosome 12, were tested for their linkage to leaf blast resistance locus using recessive class approach (RCA)50. Since the initial linkage analysis indicated STS-5 and RM1261 to be the flanking markers for the resistance locus, six additional polymorphic SSR markers viz., RM3246, RRRS12, RRS77, RRS16, RRS19 and RRS69 were selected from the genomic region bracketed by these markers for chromosome walking to the resistance gene. These internal markers were used for the genotyping of the recombinants detected with STS-5 and RM1261 to delimit the genomic region for the resistance locus. The primer sequences for the markers used in mapping were adapted from the web site of the International Rice Microsatellite Initiative (IRMI; http://www.gramene.org) and Kumar et al.33.
DNA amplification was carried out in a 12.5 µl reaction volume containing 20 ng template DNA, 0.2 mm of each dNTP, 0.2 µM of each primer, 1.5 mm MgCl2, 1X PCR buffer (10 mm Tris–HCl, 50 mm KCl, pH 8.3) and 1 unit Taq polymerase (Gotaq® DNA Polymerase, Promega). PCR amplification was carried out in a thermocycler (Proflex PCR System; Applied Biosystems, Life Technologies, USA) using the following temperature profile: initial denaturation at 94 °C for 5 min followed by 39 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s and a final extension at 72 °C for 5 min followed by rapid cooling to 4 °C. The PCR products were resolved in 4% agarose gel and visualized by ethidium bromide staining. The intensity of linkage between the markers and resistance gene(s) was deduced by the segregation analysis of 220 susceptible F2 plants. Linkage analysis was performed with Map Disto Version 2.0 software52 and the recombination frequency was converted to map distance expressed in cm (centi Morgan) using the Kosambi function53.
Delineation of the physical position of the blast resistance locus and identification of candidate resistance gene(s)
The physical map of the resistance locus was constructed in silico by aligning the sequences of flanking markers on the genome sequence of cv. Nipponbare using BLAST programme of Rice Annotation Project Database (RAP-DB) (http://rapdb.dna.affrc.go.jp/). The gene content in the target region was deduced by searching the TIGR Rice Genome Annotation Project database of cv. Nipponbare (http://rice.plantbiology.msu.edu). The gene(s) encoding nucleotide-binding site and leucine-rich repeat (NBS-LRR) proteins were shortlisted as likely candidates for the blast resistance gene identified from RIL4. The functional support for the identified candidate genes was ascertained by searching for the presence of matching full length c-DNA (FL-cDNA) transcripts and proteins in the TIGR and the Rice Annotation Project databases (http://rapdb.dna.affrc.go.jp/; http://rice.plantbiology.msu.edu).
Allelic tests with Pita 2 and Pi67 resistance genes
The allelic relationship of the leaf blast resistance gene identified from RIL4 with blast resistance genes Pita2 and Pi67 was studied by crossing the line RIL4 with genotypes IRBLta2-Pi and TDH251 harboring these genes29,54. The seed of line IRBLta2-Pi was kindly provided by Dr. N. Kobayashi, International Rice Research Institute, Philippines, while for TDH251, the seed available in our department was used. The F2 progenies of crosses RIL4 x IRBLta2-Pi and RIL4 x TDH251 along with parental genotypes were inoculated with the blast isolate, PO-HPU2216-5–2. The procedure for disease inoculation and scoring was essentially similar to that described in the preceding sections. The observed ratio of Resistant : Susceptible plants in each cross was tested for goodness of fit to 15:1 ratio excepted for duplicate independently segregating dominant genes using Chi-square test.
Ethical declarations
This study does not involve any ethical issues. There were no human or animal subjects involved in the study.
Statement regarding plant materials
The monogenic lines including IRBLta2-Pi have been procured from IRRI, Philippines under material transfer agreement. Further, all plant experiments involved in this study have been conducted in accordance to relevant regulations and guidelines.
Data availability
All the data used in the present study are included in this manuscript and supplementary information files. The genomic resources of Oryza sativa ssp. japonica cv. Nipponbare used for the construction of physical map, searching gene content and full length Transcripts (Fl-cDNA) and proteins sequences are available in public domain at http://rapdb.dna.affrc.go.jp and http://rice.plantbiology.msu.edu.
References
Tieyan, L. & Mingsheng, C. Genome evolution of Oryza. Biodivers. Sci. 22(1), 51–65 (2014).
Kumari, M. et al. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad-spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 36(11), 1747–1755 (2017).
Shim, K. S. et al. Identification of fungal (Magnaporthe grisea) stress-induced genes in wild rice (Oryza minuta). Plant Cell Rep. 22, 599–607 (2004).
Food and Agriculture Organization of the United Nations (FAOSTAT). https://www.fao.org/faostat/en/#data Accessed on July 2023
Khush, G. S. Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant Breed. 132(5), 433–436 (2013).
Khush, G. S. & Jena, K. K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In Advances in genetics, genomics and control of rice blast disease (eds Xiaofan, W. & Valent, B.) (Springer, 2009).
Pennisi, E. Armed and dangerous. Science 327(5967), 804–805 (2010).
Puri, K. D., Shrestha, S. M., Chhetri, B. K. G. & Joshi, K. D. Leaf and neck blast resistance reaction in Tropical rice lines under greenhouse condition. Euphytica 165(3), 523–532 (2009).
Wang, Z. X. et al. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 19, 55–64 (1999).
Fukuoka, S. & Okuno, K. QTL analysis and mapping of pi21 a recessive gene for field resistance to rice blast in Japanese upland rice. Theor. Appl. Genet. 103, 85–190 (2001).
Zhou, B. et al. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant. Microbe Interact. 19, 1216–1228 (2006).
Liu, X., Lin, F., Wang, L. & Pan, Q. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 176, 2541–2549 (2007).
Zhai, C. et al. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189, 321–334 (2011).
Zhao, H. et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 2039. https://doi.org/10.1038/s41467-018-04369-4 (2018).
Hayashi, N. et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510 (2010).
Chen, J. et al. A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae. J. Genet. Genomics 38, 209–216 (2011).
Ma, J. et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microb. Interact. 28(5), 558–568 (2015).
Sirithunya, P. et al. Quantitative trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa). DNA Res. 9(3), 79–88 (2002).
Ishihara, T. et al. Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice 7(1), 1–11 (2014).
Padmanabhan, S. Y., Chakrabarti, S. C., Mathur, S. C. & Veeraraghavan, J. Identification of pathogenic races of Pyricularia oryzae in India. Phytopathology 60, 1574–1577 (1970).
Wang, L. et al. Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. USA 116(37), 18479–18487 (2019).
Rathour, R., Katoch, A., Kusum, K. R. & Sharma, T. R. Virulence analysis of Magnaporthe oryzae for resistance gene deployment in north-western Himalayas. Plant Dis. Res. 26(2), 183 (2011).
Jia, Y., Zhou, E., Lee, S. & Bianco, T. Co-evolutionary Dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pi-ta-1. Phytopathology 106(7), 676–683 (2016).
Kiyosawa, S. Studies on inheritance of resistance of rice varieties to blast 3 inheritance of resistance of a rice variety Pi No. 1 to the blast fungus. Jpn. J. Breed. 16, 243–250 (1966).
Kiyosawa, S. Inheritance of resistance of the rice variety Pi No. 4 to blast. Jpn. J. Breed. 17, 165–172 (1967).
Mackill, D. J. & Bonman, J. M. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82(7), 746–749 (1992).
Inukai, T. et al. Allelism of blast-resistance genes in near isogenic lines of rice. Phytopathology 84, 1278–1283 (1994).
Sharma, T. R. et al. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genomics 274, 569–578 (2005).
Joshi, S., Dhatwalia, S., Kaachra, A., Sharma, K. D. & Rathour, R. Genetic and physical mapping of a new rice blast resistance specificity Pi-67 from a broad spectrum resistant genotype Tetep. Euphytica 215(1), 9 (2019).
Kalia, S. Genetics and mapping of neck blast resistance gene(s) from RIL4 a recombinant inbred line derivative of the broad spectrum resistant genotype Tetep. PhD. Thesis, CSKHPKV, Palampur, India (2019)
Chen, S. et al. Resistance spectrum assay and fine mapping of the blast resistance gene from a rice experimental line, IRBLta2-Re. Euphytica 195, 209–216 (2014).
Jia, Y. & Martin, R. Identification of a new locus, Ptr(t), required for rice blast resistance gene Pi-ta-mediated resistance. Mol. Plant Microb. Interact. 21(4), 396–403 (2008).
Kumar, P. et al. Genetic and physical mapping of blast resistance gene Pi-42(t) on the short arm of rice chromosome 12. Mol. Breed. 25, 217–228 (2010).
Meng, X. et al. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice 13, 19. https://doi.org/10.1186/s12284-020-00377-5 (2020).
Rybka, K., Miyamoto, M., Ando, I., Saito, A. & Kawasaki, S. High resolution mapping of the indica-derived rice blast resistance genes II. Pi-ta2 and Pi-ta and a consideration of their origin. Mol. Plant Microb. Interact. 10, 517–524 (1997).
Zhuang, J. Y. et al. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 128(3), 363–370 (2002).
Noenplab, A. et al. QTL mapping for leaf and neck blast resistance in KhaoDawk Mali 105 and JaoHom Nin recombinant inbred lines. Sci. Asia 32, 133–142 (2006).
Bryan, G. T. et al. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045 (2000).
Kalia, S. & Rathour, R. Current status on mapping of genes for resistance to leaf-and neck-blast disease in rice. 3 Biotech 9(6), 209. https://doi.org/10.1007/s13205-019-1738-0 (2019).
Dong, L. et al. Fine mapping of Pi57(t) conferring broad spectrum resistance against Magnaporthe oryzae in introgression line IL-E1454 derived from Oryza longistaminata. PloS One 12(10), e0186201. https://doi.org/10.1371/journal.pone.0186201 (2017).
Zhao, H. et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9(1), 1–12 (2018).
Okuyama, Y. et al. A multifaceted genomics approach allows the isolation of the rice Pia blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479 (2011).
Lee, S. K. et al. Rice Pi5 mediated resistance to Magnaporthe oryzae requires the presence of two nucleotide-binding-leucine-rich-repeat genes. Genetics 181, 1627–1638 (2009).
Ashikawa, I. et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pik-m specific rice blast resistance. Genetics 180, 2267–2276 (2008).
Cesari, S. et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25(4), 1463–1481 (2013).
Białas, A. et al. Lessons in effector and NLR biology of plant-microbe systems. Mol. Plant Microb. Interact. 31(1), 34–45 (2018).
Wu, C. H. et al. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 114(30), 8113–8118 (2017).
Rathour, R., Singh, B. M. & Sharma, T. R. Population structure of Magnaporthe grisea from north western Himalayas and its implications for blast resistance breeding of rice. J. Phytopathol. 152, 304–312 (2004).
Mathur, R. S., Barnett, H. L. & Lilly, V. G. Sporulation of Colletotrichum lindemuthianum in culture. Phytopathology 40, 104–114 (1950).
Zhang, Q. et al. Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc. Natl. Acad. Sci. USA 91, 8675–8679 (1994).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 8, 4321–4325 (1980).
Lorieux, M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235 (2012).
Kosambi, D. D. The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175 (1944).
Tsunematsu, H. et al. Development of monogenic lines of rice for blast resistance. Breed. Sci. 50, 229–234 (2000).
Acknowledgements
The work was supported by Indian Council of Agriculture Research under project “Molecular genetic analysis of resistance/tolerance to different stresses in rice, wheat chickpea and mustard including sheath blight complex genomics”.
Author information
Authors and Affiliations
Contributions
R.R., S.G.K., A.K.S. and T.R.S. conceived the work, and designed experiments. B.B. contributed in blast phenotyping and preparing rough draft of the manuscript. K.T. and T.D.P. performed genotyping work. KDS contributed in editing and improvement of the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Biswas, B., Thakur, K., Pote, T.D. et al. Genetic and molecular analysis of leaf blast resistance in Tetep derived line RIL4 and its relationship to genes at Pita/Pita2 locus. Sci Rep 13, 18683 (2023). https://doi.org/10.1038/s41598-023-46070-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46070-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.