Abstract
BRD4 contains two tandem bromodomains (BD1 and BD2) that recognize acetylated lysine for epigenetic reading, and these bromodomains are promising therapeutic targets for treating various diseases, including cancers. BRD4 is a well-studied target, and many chemical scaffolds for inhibitors have been developed. Research on the development of BRD4 inhibitors against various diseases is actively being conducted. Herein, we propose a series of [1,2,4]triazolo[4,3-b]pyridazine derivatives as bromodomain inhibitors with micromolar IC50 values. We characterized the binding modes by determining the crystal structures of BD1 in complex with four selected inhibitors. Compounds containing [1,2,4] triazolo[4,3-b]pyridazine derivatives offer promising starting molecules for designing potent BRD4 BD inhibitors.
Similar content being viewed by others
Introduction
The bromodomain and extraterminal (BET) protein families (BRD2, BRD3, BRD4, and BRDT) are proteins with two canonical tandem bromodomains (BD1 and BD2) that recognize acetylated lysine (Kac) at their N termini1,2. Until now, the molecular functions of these tandem domains were suggested to be transcriptional1,3 and nontranscription-related functions4,5,6,7. These two bromodomains share high sequence and structure similarity8,9. However, slight differences in surface charges and structure in the Kac binding site result in a distinct selectivity in the binding of acetylated histone/nonhistone peptides and induce various biological functions1,2.
Among BET family members, bromodomain-containing protein 4 (BRD4) has been extensively studied. As a key components of a transcriptional complex in epigenetic events, BRD4 functions as a reader, and two bromodomains of BRD4 specifically interact with Kac containing the N-terminus of histones1. The hydrophobic cavity, which is also called the Kac binding pocket4,10, possesses a functional surface that specifically interacts with Kac and has roles in epigenetic readers by forming a transcriptional multiprotein complex1,8,11. BRD4 is widely expressed in the body and is a promising therapeutic target for treating cancers10,12,13,14,15 and noncancer diseases16,17, considering its disease-related changes in expression level18,19,20.
In 2010, two BRD4 inhibitors were introduced, JQ1( +) (hereafter, JQ1) and GSK525762A (I-BET762), which have similar core chemical scaffolds3,21. After these discoveries, extensive efforts to discover BRD4 BD1/BD2 inhibitors with high efficacy and specificity have been conducted. Recently, the development of small molecule inhibitors of BRD4 has involved the following main strategies: blockade molecules (selective BET inhibitor; BETi2,22,23 and pan BET inhibitor; pan BETi24) and degraders (degrader BET; dBET25,26). Many reports have shown that pan BETis, such as JQ1 and iBET762, exhibited no selectivity between BD1 and BD2, but BD1-selective (GSK778) or BD2-selective (GSK046 and ABBV-744) BETis showed significant IC50 value differences between BD1 and BD22,9,22,23. In addition, recent studies have shown that selective targeting of BD1 and BD2 of BET proteins results in different treatment effectiveness. For example, whereas a BD1-selective inhibitor (GSK778) showed similar phenocopies of pan BETis in cancers, a BD2-selective inhibitor (GSK046) showed better effectiveness in inflammatory and autoimmune diseases2. Another report showed that BD2-selective BET family inhibitors exhibited good efficacies in treating prostate cancer22. Furthermore, targeted protein degraders or proteolysis-targeting chimeras (PROTACs; dBET1 and ARV-771) utilizing small molecules of BRD4 BETis (JQ1, I-BET762) have shown promising treatment efficacies in many types of cancers and immune-related diseases25,26. Chemical structures of representative examples of each type of inhibitors are shown in Supplementary Fig. S1.
Extensive structural studies of BRD4 bromodomains in complex with Kac peptides or various inhibitors have revealed critical residues of BD1/BD2 that participate in protein or ligand binding3,8,27,28,29. They also proposed the binding modes of ligands and contributed to structure-based inhibitor discoveries.
In this study, we present a chemical scaffold of [1,2,4]triazolo[4,3-b]pyridazine derivatives as bromodomain inhibitors. As an extension of previous works30,31, we discovered a hit molecule by the AlphaScreen assay for determining biological activity. More than thirty derivatives of the hit compound were assessed by in vitro assay, and several inhibitors with micromolar IC50 values were found. In addition, we comprehensibly analyzed our BD1–inhibitor complex structures and other ligand-bound BD1 structures deposited in the Protein Data Bank (PDB). Our results provide structural insight into the development of efficient inhibitors of BRD4 BD1/BD2 through structure-guided strategies.
Results and discussion
Analysis of the ligand binding cavity with BRD4 BD1 structures
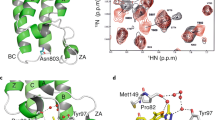
We comprehensively compared deposited structures in the PDB and determined the key ligand interacting residues by using the Protein‒Ligand Interaction Profiler (PLIP) server, which detect and list the various types of interactions between protein and ligand32,33. BD1 has four α helices (αZ, αA, αB, and αC) as the skeletal backbone, and they are linked with three loops named the ZA, AB, and BC loops (Fig. 1a). The Kac and inhibitor binding site (ligand binding site) of BD1 is formed by ZA and BC loops, and most interacting residues reside on those loops (yellow and magenta in Fig. 1a,b; the key residues are illustrated as sticks). A characteristic WPF motif (81WPF83), which is important for ligand or peptide binding, resides in the ZA loop (Fig. 1).
The overall structure of BRD4 BD1. 6KEI is aristoyagonine derivative-bound BRD4 BD1, and 2OSS is apo-BRD4 BD1. (a) Each secondary structure element is labeled in the figure. The ligand binding site of BD1 was illustrated by the cavity mode suite using the program PyMOL (Schrödinger, New York, NY, USA). In the ZA loop (yellow), three helices are illustrated in magenta. The compound (pink) was bound in the cavity. (b) Key residues participating in Kac peptide or inhibitor binding are depicted in the apo form of BRD4 BD1. Most of the key residues are positioned at the ZA loop and BC loop (blue: common hydrogen bond-forming residue, yellow: common hydrophobic interacting residues, cyan: additional ligandable residues in previous reports30,31). The figures were drawn using the program PyMOL version 2.0 (Schrödinger, New York, NY, USA, https://www.schrodinger.com/products/pymol).
Crystal structures of the substrate histone peptide-bound BD1 showed structural insight for binding drugs (Supplementary Fig. S2). In the monoacetylated lysine (mo-Kac)-bound BD1 structure (PDB code: 3JVK), the long sidechain of the acetylated lysine of the Kac peptide mostly inserted into the deep cavity and interacted with the protein through hydrophobic interactions (Supplementary Fig. S2a). In particular, the terminal acetyl group is critical for interaction. As readers of lysine acetylation, the di-acetylated lysine (di-Kac) histone tail exhibits a higher affinity than that of mo-Kac, and the di-Kac peptide-bound cocrystal structures (PDB code: 3UVW, 3UVX, and 3UVY) are also informative in characterizing the ligand binding mode in BRD4 BD11,8. The first Kac (Kac1) in the di-Kac peptide showed a similar binding mode to the mo-Kac peptide, but additional interactions can strengthen the interaction (Supplementary Fig. S2b–g). In detail, the Kac1 formed a hydrogen bond between the carbonyl of the acetyl group of Kac and the amine of Asn140 in the BC loop, and multiple hydrophobic interactions with some hydrophobic residues (Phe83, Val87, Leu92, and/or Leu94) were also found (Supplementary Fig. S2b,d,f). The second Kac (Kac2) usually forms extensive hydrophobic interactions with residues in the ZA loop, particularly, WPF motif. In some structures, Kac2 also make interactions with Asp145 and Ile146 (Supplementary Fig. S2d,f). Additionally, the other parts of the di-Kac peptides, including mainchain, forms multiple hydrogen bonds with residues in the BC loop (Supplementary Fig. S2c,e,g). It is interesting that the two individual Kac (Kac1, Kac2) residues are usually connected by several conserved water molecules near the WPF motif (Supplementary Fig. S3).
In the ZA loop, there are three 310 helices that can easily be unwound (Fig. 1). Two of them contain many ligand-interacting residues. The first turn is the conserved WPF motif, which contributes hydrophobic interactions with other ligands. The second helix possesses another conserved sequence, which contains two conserved leucine residues (92LxL94; for the x, Asn in BD1, Gly in BD2). These two leucine residues also participate in hydrophobic interactions with ligands. Taken together, the results indicate that hydrophobic residues in the WPF and LxL motifs form hydrophobic interactions with the substrates and inhibitors. In addition to these common key residues, the Val87, Tyr139, Asp145, and Ile146 residues also participate in forming hydrophobic interactions with ligands in BD1. Asn140 is an important residue that forms a hydrogen bond with the ligand. Additionally, in our recent reports, benzo-oxepinoindol derivative- and pyridin-benzotriazol derivative-BRD4 BD1 cocrystal structures revealed that Gln85, Asp88, and Tyr97 interact with the ligand and contribute to the enhancement in ligandability (Fig. 1b, illustrated in aquamarine sticks)30,31.
In BD1, there are characteristic conserved water molecules15 near the WPF motif (Supplementary Table S1). The water molecules located at conserved positions play critical roles in binding the Kac peptide and maintaining the BD1 structure by forming hydrogen bonds (Supplementary Fig. S3a,b; W1, W2 (2–1, 2–2, 2–3), W3 and W4). Three of them (W1, W3, and W4) existed in highly conserved positions and W2 molecules existed in relatively variable positions in many Kac peptide-bound BD1 structures. In detail, the W1 is the essential water for maintaining the cavity structure by bridging the mainchain carbonyl group of Pro82 and Gln85 and the W4 molecule connects two sidechains of Gln84 and Gln85, respectively (Supplementary Fig. S3). The W2 (including 2–1, 2–2, 2–3) and W3 waters usually generate additional interactions between BD1 and Kac peptides to enhance the Kac peptide-BD1 interactions. Particularly, the W3 involved in multiple interaction between mo-Kac or di-Kac moiety and Pro82. Most of the peptide-bound BD1 structures have these four conserved water molecules and many pan BETi-bound BD1 structures have W1 and W4 molecules. These suggest that disruption of waters (W1 and W4) which involved in shaping of the Kac peptide binding pocket likely unfavorable.
Although these waters are not strictly conserved in ligand-bound BD1 structures, some of them are also found in many inhibitor-bound BD1 structures (Supplementary Fig. S3c). Usually, the W3 water molecules are occupied by inhibitor molecules. Moreover, an inhibitor-bound BD1 structure (PDB code: 7W3D) do not have any four conserved waters. The sulfonamide moiety of N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine displaced W2 and other Kac binding pocket shaping waters were not also observed (Supplementary Fig. S3d)30.
Biological activity studies of [1,2,4]triazolo[4,3-b]pyridazine derivatives
To find the hit molecules for BRD4 bromodomain inhibitor discovery, we performed an in vitro AlphaScreen assay with in-house chemical libraries. The hit molecule and its derivatives were tested to measure the IC50 values against BD1 (Fig. 2, Supplementary Table S2). In the first-line screening, we found the compound 5 as an initial hit (Supplementary Table S2), which contains [1,2,4]triazolo[4,3-b]pyridazine as the chemical core scaffold.
Selected [1,2,4]triazolo[4,3-b]pyridazine derivatives for BETi. (a) The core scaffold chemical structure for derivative studies and four selected compounds for complex structure determination. (b) Representative results for the in vitro inhibition test by AlphaScreen (PerkinElmer, Waltham, MA, USA).
To examine the inhibitory activity according to the chemical structure variations, we replaced the chemical groups at the R1 and R2 sites of the core scaffold and monitored inhibitory activities (Supplementary Table S2). The in vitro tests showed that inhibitory effects were not greatly improved when various indole-based groups were introduced to the R2 site and various chemical groups were introduced at the R1 site (5–12). In detail, modification of the C5 position of indole with a fluoro group (7) or methoxy group (8) showed the nearly same blocking efficacy (carbon position numbers are indicated in Fig. 2). In addition, the substitution from methyl to trifluoromethyl at R1 did not much improved the IC50 value (6). Other modifications were conducted at the C2 and C3 positions in N-methylpropan-1-amine, a linker between the core scaffold and indole. The trifluoromethyl moiety at R1 and aryl addition at C3 in linker (9) increased the IC50 value by only twofold compared to that of 6, which suggests that the addition of an aryl moiety on the ethylene linker showed no positive effect on BD1 binding. Furthermore, changes from methyl to ethyl at R1 and methanol addition at the C2 position in the N-methylpropan-1-amine linker (12) showed decreased blocking efficiency compared to that of 5. The inhibitory efficacy was worse (no inhibition) with the bulky ortho fluorobenzyl substitution at R1 and the formation of a piperidine ring between indole and [1,2,4]triazolo[4,3-b]pyridazine (11). Methylcyclopropane substitution at R1 and chloro- and fluoro- ortho-substitution at the C5 and C6 positions of indole showed no inhibitory efficiency in the measured range (10).
A series of chemical optimization trials by changing the R2 with piperidine derivatives (13–27) provided very diverse results. While substitution of only the piperidine group at R1 (13) showed no blocking activities, but surprisingly, introducing 4-methylpiperidine (14) restored inhibitory efficacy. Next, we further examined an additional thirteen piperidine-based derivatives (15–27). N,N-dimethylmethane sulfonamide substitution at R1 with compound 14 (15) and oxygen and sulfur substitution at the C4 position of piperidine (16, 17, 18) showed no inhibitory efficacy. Only two derivatives (19, 21) exhibited inhibitory activities for spiro ring compound substitution at the C4 position of piperidine (19–23). Additionally, R1 substitution with hydrophobic chains and cyclic rings as well as R2 substitution at the C4 position of piperidine showed no inhibitory activities (24–27). When linear chains were substituted at R2 (28–34), only an ethylpropanoate-containing derivative (33) showed inhibitory activity, suggesting difficulties in BD1 binding with these derivatives. Substitution of the benzyl group instead of the indole group (35–37) resulted in similar or slightly improved activities. However, benzyl groups with longer amide-based linkers (38–40) showed no inhibitory activities.
We also tested inhibitory activities with four selected hit compounds (compounds 5, 6, 12, and 14) for BD2 and found that these compounds also have inhibitory activities for BD2, suggesting the pan BETis (Fig. 2b; Table 1).
Structural studies of BD1:[1,2,4]triazolo[4,3-b]pyridazine derivatives
To explain the rationales of the inhibitory activities of the derivatives, we tried to determine the crystal structure of BD1 in complex with inhibitors. We successfully obtained four crystal structures of BD1 in complex with [1,2,4]triazolo[4,3-b]pyridazine derivatives (compounds 5, 6, 12, and 14; Figs. 2 and 3). All the crystal structures were high-resolution structures (1.4 − 1.53 Å; Supplementary Table S3, Supplementary Fig. S4).
Cocrystal structure of each compound in BRD4 BD1. (a–d) The compound 5, 6, 12, 14-bound BD1 structures. Each interaction is illustrated by dashed lines (hydrogen bond (blue dash), π–π stacking interaction (cyan dash), hydrophobic interaction (yellow dash), conserved water molecule-mediated interactions (green dash)). (e) Superposition of four ligand-bound BD1 structures. The protein–ligand interactions were analyzed and quantified using PLIP servery32,33. The figures were drawn using the program PyMOL version 2.0 (Schrödinger, New York, NY, USA).
Most of the published Kac mimetic BRD4 BD1 inhibitor-bound structures showed that their interactions use conserved key interacting residues and water molecules. Our [1,2,4]triazolo[4,3-b]pyridazine derivative structures showed similar binding modes (Fig. 3). The overall orientations of [1,2,4]triazolo[4,3-b]pyridazine are nearly the same in all four compounds. The conserved WPF motif interacts with chemical groups at the R1 site and indole (or methylpiperidine) groups at the R2 site of the hit (Fig. 3).
Among many interacting key residues that identified by PLIP server analyses, each derivative utilizes approximately five to ten residues for interactions. In detail, chemical groups at the C3 position of the core scaffold methyl (5, 14), trifluoromethyl (6), and ethyl (12) toward the WPF motif in the Kac binding pocket and indole or methylpiperidine ring are exposed to the solvent region but also toward the WPF motif. The N1 nitrogen atom of the triazole of the core scaffold forms hydrogen bonds with Asn140. Two hydrophobic interactions between the core scaffold and Leu94 and the core scaffold and Tyr139 are common features in 5, 6, and 12. These hydrophobic interactions are not found in 14 (Fig. 3d). In addition, the hydrophilic trifluoromethyl moiety in 6 lost its hydrophobic interaction with Val87, while others maintained this interaction. The triazole moiety of 5 was pulled to Asn140, and it generated π-stacking interactions with Tyr97 as well as water-mediated hydrogen bond interactions (Fig. 3a). A superposition of all four structures showed slight positional movement of the core scaffold in the 5-, 6-, 12-, and 14-bound BD1 structures (Fig. 3e). There is no large conformational change in BRD4 BD1 in the [1,2,4]triazolo[4,3-b]-BRD4 BD1 complex structures, and ligand recognition is likely governed by conformational selection rather than induced fit34.
Another common feature of the compound 5-, 6-, 12-BD1 complex is that the indole group contacts Trp81, Asp145, and Ile146 through hydrophobic interactions (Fig. 3a–c). On the other hand, there are also differences between the three complex structures for adapting conformational selection; the indole group of 12 is more bent and does not form hydrophobic interactions with Pro82. The compound 14 contains a piperidine group at the R2 position of the scaffold instead of indole, which does not interact with Asp145 and Ile146 to form hydrophobic interactions but still interacts with Trp81 and Pro82 (Fig. 3d).
The conserved water molecules in the ligand binding site play different roles according to inhibitor type. The W1 molecule in the 6-BD1 complex contributed to an additional hydrogen bond with the trifluoromethyl moiety (Fig. 3b). The hydroxyl group in 12 adopted an additional water molecule (W2-1, W2-3, and W5)-mediated interaction for ligand binding (Fig. 3c). The compound 14 showed a retained conserved water with the WPF motif, but these water molecules did not participate in water-mediated ligand binding (Fig. 3d). Only 5 contains W3, similar to Kac peptide, and it interacts with N5 of the imine group at [1,2,4]triazolo[4,3-b]pyridazine core scaffold, forming a water-mediated interaction (Fig. 3a). The 12-BD1 complex provides insight for drug design because the ethyl moiety of R1 was bent deep inside the cavity due to its hydrophobicity (Fig. 3c). It could also be inferred from our structures that hydrophobic R1 substitutions, such as methylcyclopropane, methylcyclobutane, and isobutane (10, 24–27), may not be capable of displacing W1 and W2 waters to bind and inhibit BD1 (Fig. 3).
Bromodomains are structurally conserved domains in a broad range of proteins, but they have unique and distinct ligands. It has been suggested that a variety of bromodomain-containing proteins exhibit sequence selectivity for Kac peptides of histone and nonhistone proteins due to the electrostatic properties of the cavity1,8. Electrostatic complementarity assessment between protein and ligand surfaces is an important consideration for drug discovery35. In this manner, we created the electrostatic potential (ESP) surface of each compound, including the potent pan BETi JQ1, with the ESP surface visualization suite in Avogadro36 (Fig. 4a). Interestingly, the charged regions of the compounds and the surface of the BD1 ligand binding cavity were electrostatically complementary (Fig. 4). Because diazene N1 and N2 were relatively negatively charged, the C7 and C8 positions of [1,2,4]triazolo[4,3-b]pyridazine showed a relatively positive charge and were positioned near Asn140 (Fig. 4b, region I), which is the negatively charged surface. The C4 and C5 positions in indole and the C2 and C3 positions in piperidine were electrically neutral, and they interacted with Trp81, which is an electrically neutral residue (Fig. 4b, region III). It is an exception that the fluorine atoms of the trifluoromethyl moiety (6), which are highly negative, were close to electrically neutral region II. However, the fluorine atom of 6 did not directly contact neutral region II but formed water-mediated interactions (Fig. 3). The compound 14, the smallest compound in this study, showed that the surfaces of the compound and binding position of BD1 were electrostatically complementary as well. As shown in Fig. 4b, JQ1 represents a strongly electrical complement surface with BRD4 BD1. The C6 and C6a positions of thienotriazolodiazepines are strongly positively charged and interact with region I. In addition, the dimethylthiophene of JQ1 is neutral and faces region II. Similar to our structure, it interacted with region II through water-mediated interactions. However, the W3 position was occupied by a dimethylthiophene group, and the sulfur atom formed a hydrogen bond with W2 (Supplementary Fig. S3c, magenta dashes)37,38.
Illustration of the electrostatic surface of each compound and BRD4 BD1. (a) The electrostatic surface of [1,2,4]triazolo[4,3-b]pyridazine derivatives. (b) The electrostatic potential surface of BRD4 BD1. The figures were drawn using the program Avogadro version 1.2.0 (http://avogadro.cc/) PyMOL version 2.0 (Schrödinger, New York, NY, USA).
Conclusions
Overall, we found a series of [1,2,4]triazolo[4,3-b]pyridazine derivatives as pan BETi scaffolds. As a structural activity relationship study, we studied four groups of chemical derivatives in R2, which were indole-, aryl-, aliphatic-, and piperidine-based moieties. Additionally, a small change in R1 was also conducted at the same time. The study showed that a series of [1,2,4]triazolo[4,3-b]pyridazine derivatives showed micromolar IC50 values in BRD4 BD1 inhibition. The actual binding characteristics of the four compounds could be explained by X-ray cocrystal structural studies. All four compound-bound structures showed that BD1 residues, which are already known to be important for ligand binding, also participated in the four ligand interactions. In addition, the conserved water molecules, shown in many previous structure reports, also conserved our inhibitor-bound BD1 structures, contributing to ligand binding. Finally, the electrostatic potential surface of each inhibitor and BRD4 BD1 were studied, and ligand interactions could be explained. Through these studies, we proposed [1,2,4]triazolo[4,3-b]pyridazine derivatives as starting points for the structure-based discovery of BETis.
Methods
BRD4 BD1 expression and purification
BRD4 bromodomain 1 (BD1; 44 − 168) protein expression and purification were performed according to a previous method. The BRD4 BD1 plasmid, subcloned into a modified pET28b vector (TEV cleavage site inserted at N-terminal; BamHI, XhoI), was transformed into BL21(DE3) and cultured with Terrific Broth. After 0.1 mM isopropyl-D-thiogalactoside induction when the OD600 reached approximately 0.8, the cells were incubated with vigorous shaking at 18 °C overnight. After harvest, the cells were resuspended, and recombinant BRD4 BD1 was purified with Ni–NTA resin. To cleave the His6x tag, 2.5 mg of TEV protease was added to 100 mg of purified BRD4 BD1. Further purification was performed with Ni–NTA resin to collect the cleaved BRD4 BD1 with the flowthrough fraction, and size-exclusive chromatography was performed to increase the purity of the recombinant protein. BRD4 BD1 was concentrated to 12 mg/mL, which was measured by nanodrop, converted with the calculated extinction coefficient, and stored at −80 °C until crystallization.
AlphaScreen enzyme assay
The AlphaScreen assay was described previously and performed according to the manufacturer’s protocol (PerkinElmer, USA). The reaction buffer was 50 mM HEPES pH 7.4, 100 mM NaCl, 0.1% BSA, and 0.05% CHAPS. The whole reaction and screening were performed using OptiPlateTM-384 (PerkinElmer, Waltham, MA, USA).
The compounds, acetylated peptide [SGRGK(Ac)GGK(Ac)GLGK(Ac)GGAK(Ac)RHRK-biotin] and purified bromodomain protein were added to the OptiPlateTM-384 plate. Then, streptavidin-coated donor beads and anti-GST AlphaScreening acceptor beads were added. The incubation condition was 25 °C for 1 h using a Thermomixer C (Eppendorf, USA). The blocking efficacy (IC50) values were obtained by 8-point titration (from 0.0096 to 32 μM). The IC50 values of inhibitors were evaluated with GraphPad Prism 7 using normalized and inhibition analysis suites.
Chemistry
Most of the compounds were synthesized by the following procedure in Scheme 1 (detailed description are in Chemistry experimental section in Supplementary materials S1). Unless otherwise stated, all reactions were performed under an inert (N2) atmosphere. Reagents and solvents were of reagent grade and purchased from Sigma‒Aldrich, Alfa Aesar, and TCI Tokyo. Flash column chromatography was performed using silica gel 60 (230–400 mesh, Merck) with the indicated solvents. Thin-layer chromatography was performed using 0.25 mm silica gel plates. Proton nuclear magnetic resonance (1H NMR) spectra were recorded on a BRUKER ultrashield 300 MHz NMR spectrometer at 25 °C. Chemical shifts are reported in parts per million (ppm). 1H NMR data are reported as follows: chemical shift (δ ppm) (multiplicity, integration, coupling constant [Hz]). Multiplicities are reported as follows: s = singlet, d = doublet, t = triplet, q = quartet, and m = multiplet. The residual solvent peak was used as an internal reference. The mass spectra were obtained using Acuity™ waters A06UPD9BM and Agilent Technologies SG12109048. Prior to biological testing, the final compounds were confirmed to be > 95% pure by ultra-performance liquid chromatography (UPLC) using a Waters ACQUITY H-class system fitted with a C18 reversed-phase column (ACQUITY UPLC BEH C18: 2.1 mm × 50 mm, Part no. 186002350) according to the following conditions: (A) H2O + 0.1% formic acid, (B) CH3CN + 0.1% formic acid, (C) methanol (MeOH) + 0.1% formic acid; (Ι) a gradient of 95% A to 95% B over 5 min, (II) a gradient of 95% A to 95% C over 5 min.
Synthetic route of compounds 5, 13–23, 31–33 and 35–37a. aReagents and conditions: (i) anhydrous n-BuOH, 120 °C, 16 h; (ii) KI, conc. HCl, ethanol, 80 °C, 24 h/72 h; (iii) 4 M HCl in dioxane: (iv) p-TSA, anhydrous ethanol, 80 °C, 14 h; (v) TFA, DCM, 1 h: (vi) N,N-dimethylsulfamoyl chloride, pyridine, 0 °C to RT, 2 h.
Scheme 1 shows the chemical synthesis of compounds (5, 13–23, 31–33 and 35–37), starting with compound 1 and acetohydrazide (2), which were obtained commercially and reacted together in anhydrous n-butanol to generate cyclized compound 3. The compound 3 was further reacted with amines (a-o) using catalytic amounts of KI and conc. HCl (concentrated HCl, 12N HCl) together in ethanol inside a sealed tube at 80 °C for 24 and/or 72 h to obtain the corresponding compounds (5, 13, 14, 16–21, 31–33, 35–37). Compounds 22 and 23 were obtained by deprotecting the Boc group using 4 M HCl in dioxane.
Intermediate 4a was synthesized by reacting 1 with commercially available tert-butyl (2-hydrazineyl-2-oxoethyl)carbamate by employing condition (iv) of Scheme 1. The obtained 4a was subjected to nucleophilic substitution by 4-methylpiperidine using condition (ii) of Scheme 1 to produce intermediate 4b, which was further treated with TFA and DCM to remove the Boc group and generate intermediate 4c. Intermediate 4c was reacted with N,N-dimethylsulfamoyl chloride and base pyridine to produce compound 15. Throughout the synthesis of compounds shown in the scheme, they were obtained in reasonable yields unless otherwise stated (Scheme 2).
Compound 33 was obtained as described in Scheme 1 and then hydrolyzed by treatment with lithium hydroxide monohydrate to generate compound 28. This compound was reacted with the peptide coupling agent HATU, base TEA and aniline to generate compound 38 with a lower yield of 17% (Scheme 3).
Synthetic route of compounds 29, 34 and 38–40a. aReagent and conditions: (i) 4 M HCl in dioxane, 1 h, RT; (ii) ethanesulfonyl chloride, DIPEA, anhydrous ethanol, 0 °C to RT, 2 h; (iii) propanoic acid, HATU, TEA, anhydrous N,N-dimethylformamide, 2 h, RT; (iv) ethyl isocyanate, DIPEA, acetonitrile, 5 h, RT; (v) benzoyl chloride, pyridine, 0 °C to RT, 4 h; (vi) phenyl isocyanate, DIPEA, acetonitrile, RT, 5 h.
Compound 32 from Scheme 1 was treated with 4 M HCl in dioxane to achieve intermediate 32a as an amine-monohydrochloride salt, which was then further subjected to the synthesis of five different compounds (29, 34, 38–40). Compound 29 was achieved by reaction between intermediate 32a and ethanesulfonyl chloride in solvent ethanol and DIPEA as base; compound 34 was obtained by reaction among intermediate 32a, propionic acid and peptide coupling reagent HATU. Compounds 38 and 40 were obtained in a similar manner by reaction of intermediate 32a with the corresponding isocyanate reagents. Intermediate 32a, when reacted with benzoyl chloride and base pyridine, produced compound 39. The overall isolated yield was lower than that of compounds 38 and 40.
Chemicals
Some chemical derivatives of compound 5 were searched using online chemical similarity search tools (https://mcule.com/search/) and purchased for further studies. Before use, all compounds were dissolved in 100% dimethyl sulfoxide (DMSO) as stock solutions of 100 mM. The supplier and catalog number are described below.
N-[2-(1H-indol-3-yl)ethyl)-3-(trifluoromethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine (Vitas-M, STK651245) (6), N-[2-(5-fluoro-1H-indol-3-yl)ethyl]-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine (Enamine, Z1220635364) (7), N-[2-(5-methoxy-1H-indol-3-yl)ethyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine (Enamine, Z1192171101) (8), N-[2-(1H-indol-3-yl)-2-phenylethyl]-3-(trifluoromethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine (Enamine, Z219181640) (9), N-[2-(5-chloro-6-fluoro-1H-indol-3-yl)ethyl]-3-cyclopropyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine (Enamine, Z2701558508) (10), 6-Fluoro-2-[3-(2-fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-yl]-1,3,4,9-tetrahydropyrido[3,4-b]indole (Enamine, Z4500949681) (11), 2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol (Z1272770102) (12), 1-(3-Isopropyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)piperidine-4-carboxylic acid (Vitas M, STK719914) (24), N-(3-methylbutyl)-1-[3-(propan-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-6-yl]piperidine-4-carboxamide (Vitas M, STK635934) (25), N-cyclopropyl-1-[3-(propan-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-6-yl]piperidine-4-carboxamide (Vitas M, STK6497959) (26), 3-cyclobutyl-6-(4-isopropylpiperazino)[1,2,4]triazolo[4,3-b]pyridazine (ChemDiv, Z606-3990) (27).
Complex structural study of BRD4 BD1 and selected compounds
All crystals were obtained by the hanging-drop vapor-diffusion method at 14 °C, following previous protocols30,31. Before crystallization, the BRD4 BD1 protein stock was thawed slowly on ice, and the protein was diluted tenfold (1.2 mg/mL) with storage buffer (10 mM HEPES-OH pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, and 10 mM dithiothreitol). In addition, compounds were mixed with a tenfold molar ratio compared to the protein stock and incubated at 4 °C with mild rotation overnight. The next day, BRD4 BD1 and compound solutions were concentrated tenfold, and the stocks were used for vapor diffusion crystallization by hanging drop. Each protein-inhibitor mixture and reservoir solution were mixed at a 1:1 ratio, and the formulation was sodium formate (ranging from 5 to 6 M) and glycerol (ranging from 2 to 12%). Crystals can be observed within 3 days and were grown over 10 days for full growth.
The diffraction dataset was collected using either the Pohang Light Source (PLS) Beamline 11C station with a Dectris Pilatus 6 M detector or the PLS Beamline 7A station with an ADSC Quantum 270 detector (Pohang, Korea). The reflected raw dataset was scaled by HKL200039. Molecular replacement solutions were processed by the PHASER suite in the PHENIX program suite, and the BRD4 BD1 structure (PDB code: 6KEK) was used as a search model31,40,41. Real space refinement and refinement were processed using the programs COOT and PHENIX, with repeated cycles42. Coordinates and restraint files of compounds were generated by MAESTRO (Schrödinger, New York, NY, USA) and the eLBOW suite in the PHENIX program43. Electron density maps for each inhibitor are shown in Supplementary Fig. S4.
Electrostatic potential surface creation of compounds
The electrostatic potential surface creation was conducted by the “create surfaces” suit. The electrostatic potential surface creation was conducted by Avogadro version 1.2.0 (http://avogadro.cc/)36. The force field of each compound was changed as a universal force field (UFF), and then surface creation was processed. The chemical structure was extracted from each deposited PDB data to preserve the stereochemistry of the conformational selection.
Data availability
All crystallographic coordinates and structure factors were deposited in the PDB under the accession codes 8GQ0 (compound 5), 7YQ9 (compound 6), 7YMG (compound 12), and 8GPZ (compound 14).
References
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231. https://doi.org/10.1016/j.cell.2012.02.013 (2012).
Gilan, O. et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science 368, 387–394. https://doi.org/10.1126/science.aaz8455 (2020).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073. https://doi.org/10.1038/nature09504 (2010).
Donati, B., Lorenzini, E. & Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 17, 164. https://doi.org/10.1186/s12943-018-0915-9 (2018).
Barrows, J. K. et al. BRD4 promotes resection and homology-directed repair of DNA double-strand breaks. Nat. Commun. 13, 3016. https://doi.org/10.1038/s41467-022-30787-6 (2022).
Yang, M. et al. Bromodomain-containing protein 4 (BRD4) as an epigenetic regulator of fatty acid metabolism genes and ferroptosis. Cell Death Dis. 13, 912. https://doi.org/10.1038/s41419-022-05344-0 (2022).
Chen, I. P. & Ott, M. Viral hijacking of BET proteins. Viruses 14, 2274 (2022).
Filippakopoulos, P. & Knapp, S. The bromodomain interaction module. FEBS Lett. 586, 2692–2704. https://doi.org/10.1016/j.febslet.2012.04.045 (2012).
Modukuri, R. K. et al. Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections. Proc. Natl. Acad. Sci. 119, e2122506119. https://doi.org/10.1073/pnas.2122506119 (2022).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. https://doi.org/10.1038/nature10509 (2011).
Fujisawa, T. & Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246–262. https://doi.org/10.1038/nrm.2016.143 (2017).
Chen, Y.-L. et al. BRD4 inhibitor GNE987 exerts anti-cancer effects by targeting super-enhancers in neuroblastoma. Cell Biosci. 12, 33. https://doi.org/10.1186/s13578-022-00769-8 (2022).
Pang, Y. et al. The BRD4 inhibitor JQ1 suppresses tumor growth by reducing c-Myc expression in endometrial cancer. J. Transl. Med. 20, 336. https://doi.org/10.1186/s12967-022-03545-x (2022).
Zheng, Z.-Z. et al. Super-enhancer-controlled positive feedback loop BRD4/ERα–RET–ERα promotes ERα-positive breast cancer. Nucleic Acids Res. 50, 10230–10248. https://doi.org/10.1093/nar/gkac778 (2022).
Zhang, M.-F. et al. Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia. Acta Pharmacol. Sin. 43, 2735–2748. https://doi.org/10.1038/s41401-022-00881-y (2022).
Shahid, S. et al. Small molecule BRD4 inhibitors apabetalone and JQ1 rescues endothelial cells dysfunction, protects monolayer integrity and reduces midkine expression. Molecules 27, 7453 (2022).
Zhong, X. et al. JQ1 attenuates neuroinflammation by inhibiting the inflammasome-dependent canonical pyroptosis pathway in SAE. Brain Res. Bull. 189, 174–183. https://doi.org/10.1016/j.brainresbull.2022.09.007 (2022).
Lambert, J. P. et al. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol. Cell 73, 621-638.e617. https://doi.org/10.1016/j.molcel.2018.11.006 (2019).
Delmore, Jake E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. https://doi.org/10.1016/j.cell.2011.08.017 (2011).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. https://doi.org/10.1038/nature10334 (2011).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123. https://doi.org/10.1038/nature09589 (2010).
Faivre, E. J. et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578, 306–310. https://doi.org/10.1038/s41586-020-1930-8 (2020).
Sheppard, G. S. et al. Discovery of N-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain inhibitor with selectivity for the second bromodomain. J. Med. Chem. 63, 5585–5623. https://doi.org/10.1021/acs.jmedchem.0c00628 (2020).
Tsujikawa, L. M. et al. Breaking boundaries: Pan BETi disrupt 3D chromatin structure, BD2-selective BETi are strictly epigenetic transcriptional regulators. Biomed. Pharmacother. 152, 113230. https://doi.org/10.1016/j.biopha.2022.113230 (2022).
Liu, L. et al. Targeted BRD4 protein degradation by dBET1 ameliorates acute ischemic brain injury and improves functional outcomes associated with reduced neuroinflammation and oxidative stress and preservation of blood–brain barrier integrity. J. Neuroinflammation 19, 168. https://doi.org/10.1186/s12974-022-02533-8 (2022).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 7124–7129. https://doi.org/10.1073/pnas.1521738113 (2016).
Demont, E. H. et al. Fragment-based discovery of low-micromolar ATAD2 bromodomain inhibitors. J. Med. Chem. 58, 5649–5673. https://doi.org/10.1021/acs.jmedchem.5b00772 (2015).
Ember, S. W. J. et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem. Biol. 9, 1160–1171. https://doi.org/10.1021/cb500072z (2014).
Zhang, G. et al. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J. Med. Chem. 56, 9251–9264. https://doi.org/10.1021/jm401334s (2013).
Pandit, N. et al. Discovery of BET specific bromodomain inhibitors with a novel scaffold. Bioorg. Med. Chem. 72, 116967. https://doi.org/10.1016/j.bmc.2022.116967 (2022).
Yoo, M. et al. Synthesis and structure–activity relationships of aristoyagonine derivatives as Brd4 bromodomain inhibitors with X-ray co-crystal research. 26, 1686. https://doi.org/10.3390/molecules26061686 (2021).
Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F. & Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43, W443-447. https://doi.org/10.1093/nar/gkv315 (2015).
Adasme, M. F. et al. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 49, W530–W534. https://doi.org/10.1093/nar/gkab294 (2021).
Vogt, A. D. & Di Cera, E. Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902. https://doi.org/10.1021/bi3006913 (2012).
Rathi, P. C., Ludlow, R. F. & Verdonk, M. L. Practical high-quality electrostatic potential surfaces for drug discovery using a graph-convolutional deep neural network. J. Med. Chem. 63, 8778–8790. https://doi.org/10.1021/acs.jmedchem.9b01129 (2020).
Hanwell, M. D. et al. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 4, 17. https://doi.org/10.1186/1758-2946-4-17 (2012).
van Bergen, L. A. H. et al. Revisiting sulfur H-bonds in proteins: The example of peroxiredoxin AhpE. Sci. Rep. 6, 30369. https://doi.org/10.1038/srep30369 (2016).
Zhou, P., Tian, F., Lv, F. & Shang, Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins 76, 151–163. https://doi.org/10.1002/prot.22327 (2009).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. https://doi.org/10.1016/S0076-6879(97)76066-X (1997).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. https://doi.org/10.1107/S0021889807021206 (2007).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877. https://doi.org/10.1107/S2059798319011471 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501. https://doi.org/10.1107/s0907444910007493 (2010).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. Sect. D Struct. Biol. D65, 1074–1080. https://doi.org/10.1107/S0907444909029436 (2009).
Acknowledgements
We thank the beamline staff of Pohang Light Source (Beamline 7A, 11C) for their assistance during X-ray data collection. This work was supported by National Research Foundation of Korea (NRF) grants funded by the Ministry of Science and ICT (NRF-2023R1A2C1002654, NRF-2019R1A2C1002545, NRF-2018R1A5A2023127 to B.I.L., and NRF-2020M2D9A2094158 to K.-Y.J.), and a National Cancer Center research grant (2210340 and 2310200 to B.I.L.), and Korea Drug Development Fund (HN21C0226000021 to C.H.P.).
Author information
Authors and Affiliations
Contributions
J.H.K., N.P., M.Y. and T.H.P. conducted experiments and contributed to data analysis, visualization, and manuscript preparation. J.U.C. performed the experiments. J.H.K., C.H.P., K.-Y.J., and B.I.L. designed the research and wrote the manuscript with input from all coauthors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, JH., Pandit, N., Yoo, M. et al. Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure–activity relationship study. Sci Rep 13, 10805 (2023). https://doi.org/10.1038/s41598-023-37527-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-37527-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.