Abstract
APOE-ɛ4 risk on Mild Cognitive Impairment (MCI) and Alzheimer’s Disease (AD) differs between race/ethnic groups, presumably due to ancestral genomic background surrounding the APOE locus. We studied whether African and Amerindian ancestry-enriched genetic variants in the APOE region modify the effect of the APOE-ɛ4 alleles on Mild Cognitive Impairment (MCI) in Hispanics/Latinos. We defined African and Amerindian ancestry-enriched variants as those common in one Hispanic/Latino parental ancestry and rare in the other two. We identified such variants in the APOE region with a predicted moderate impact based on the SnpEff tool. We tested their interaction with APOE-ɛ4 on MCI in the Study of Latinos-Investigation of Neurocognitive Aging (SOL-INCA) population and African Americans from the Atherosclerosis Risk In Communities (ARIC) study. We identified 5 Amerindian and 14 African enriched variants with an expected moderate effect. A suggestive significant interaction (p-value = 0.01) was found for one African-enriched variant, rs8112679, located in the ZNF222 gene fourth exon. Our results suggest there are no ancestry-enriched variants with large effect sizes of interaction effects with APOE-ɛ4 on MCI in the APOE region in the Hispanic/Latino population. Further studies are needed in larger datasets to identify potential interactions with smaller effect sizes.
Similar content being viewed by others
Introduction
The APOE-ε4 allele is the strongest known genetic risk factor for MCI and Alzheimer’s Disease (AD)1. However, its effect on cognitive outcomes in Hispanics/Latinos is weaker and inconsistent compared to non-Hispanic Whites2,3. The genomes of Hispanics/Latinos are admixed, consisting of three predominant continental ancestries: Amerindian, African, and European4. In a recent publication, we performed an association study of APOE alleles and neurocognitive traits in middle-aged and older U.S. Hispanics from the SOL-INCA5. We discovered that their effects were modified by continental global genetic ancestry, e.g., Amerindian genetic ancestry protects from the risk conferred by APOE-ε4 on cognitive decline. Other studies conducted in admixed populations have also concluded that ancestry-specific genetic differences, either genome-wide or in the surrounding region of the APOE gene, modify the effect of APOE alleles on ADRD6,7,8,9. For example, a recent study explored missense variants in the APOE region and identified a variant coinherited with APOE-ε4 which mitigates the AD risk effect, and another variant coinherited with APOE-ε3, which has a protective effect10. Both variants were assessed in Europeans, due to the paucity of variant carriers in non-European ancestries. Another recent study identified a variant proximal to the APOE region, reducing the APOE-ε4 risk for AD in African Ancestry9.
We recently developed a computationally efficient method for ancestry-specific frequency estimation of bi-allelic genetic variants in a multi-way admixed population11. We published a database of ancestry-specific allele frequencies estimated from the HCHS/SOL Hispanic/Latino population. This unique dataset enables us to focus on ancestry-enriched genetic variants from African and Amerindian ancestries, that were previously understudied compared to European-enriched variants. We hypothesize that African and/or Amerindian ancestry-enriched genetic variants interact with the APOE alleles in their associations with AD and related cognitive outcomes, thus potentially explaining the modification of the global genetic ancestry on the effect of the APOE allele on cognitive outcomes5. Here, we study the modification effect of African and Amerindian ancestry-enriched genetic variants in the APOE region, on the effect of the APOE-ɛ4 alleles on MCI and MCI + , where MCI + defines a subset of the MCI group with suspected severe cognitive impairment, in the Study of Latinos-Investigation of Neurocognitive Aging (SOL-INCA) population. We focus on the enriched Amerindian and African variants in a region of 6Mbp encompassing the APOE gene. We further consider only variants with a moderate estimated effect based on SNPEff annotation. We conduct interaction analyses between the identified variants and the APOE-ε4 allele and its effect on MCI in SOL-INCA. We attempt replication of one suggestive interaction association between an African-enriched variant and APOE-ε4 allele on MCI in African Americans from the Atherosclerosis Risk In Communities (ARIC) study.
Methods
Study population
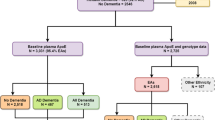
The HCHS/SOL is a population-based longitudinal multi-site cohort study of Hispanic/Latino adults in the U.S. that primarily enrolled participants from six self-identified backgrounds: Cuban, Central American, Dominican, Mexican, Puerto Rican, and South American12,13. A total of 16,415 adults, 18–74-year-old, were enrolled in the baseline visit at four field centers (Bronx, NY, Chicago, IL, Miami, FL, and San Diego, CA) (2008–2011). The baseline visit assessed cognitive function in 9,714 individuals aged 45 years or older. SOL-INCA is an ancillary study of HCHS/SOL, focusing on the middle-aged and older adult group who underwent cognitive assessment at visit 1. Overall, 6,377 individuals 50 or older with baseline cognitive testing participated in the SOL-INCA examination at or after HCHS/SOL visit 2, with an average of 7 years after the baseline exam. Of the 6,377 participants, 2140 were excluded from analyses (1701 did not consent for genetic data, 210 without APOE data, 76 had missing cognitive outcomes, and 153 had missing covariates and/or genetic variant reads), totalling an analytic sample of 4237 individuals.
The HCHS/SOL was approved by the institutional review boards (IRBs) at each field center, where all participants gave written informed consent in their preferred language (Spanish/English) to use their genetic and non-genetic data, and by the Non-Biomedical IRB at the University of North Carolina at Chapel Hill, to the HCHS/SOL Data Coordinating Center. All IRBs approving the study are: Non-Biomedical IRB at the University of North Carolina at Chapel Hill. Chapel Hill, NC; Einstein IRB at the Albert Einstein College of Medicine of Yeshiva University. Bronx, NY; IRB at Office for the Protection of Research Subjects (OPRS), University of Illinois at Chicago. Chicago, IL; Human Subject Research Office, University of Miami. Miami, FL; Institutional Review Board of San Diego State University, San Diego, CA. The study reported here was approved by the Mass General Brigham IRB under protocol #2019P000057. All methods and analyses of HCHS/SOL participants’ materials and data were carried out in accordance with human subject research guidelines and regulations.
Neurocognitive outcome
Individuals were classified with MCI according to National Institute on Aging-Alzheimer’s Association criteria based on cognitive tests and self-reports14. Details about the SOL-INCA MCI diagnostic operational procedures have been previously published15,16. MCI was defined according to three criteria that had to be satisfied: (a) for any of the cognitive tests performed at the SOL-INCA exam, the score was lower than −1 standard deviation (SD) of the mean, with means and SDs being defined based on SOL-INCA robust internal norms; (b) the rate of a global measure of yearly cognitive decline, estimated between the HCHS/SOL baseline and the SOL-INCA exam, was faster than −0.055 SD; (c) using the Everyday Cognition 12-item version (E-Cog12) questionnaire17 a participant self-reported subjective cognitive decline. The MCI group also included individuals classified as MCI + based on satisfying two conditions: (a) a cognitive test score lower than − 2 SD below the mean of any cognitive test performed at the SOL-INCA exam (with means and SDs based on SOL-INCA internal norms); (b) more than minimal impairment in self-reported instrumental activities of daily living (IADL)18. Individuals with MCI + were pooled together with the MCI individuals and together defined the MCI group.
Genetic data
APOE genotyping was performed using commercial TaqMan assays previously described19. For individuals with missing APOE genotypes, we computed the genotypes based on phased whole-genome sequencing (WGS) data from TOPMed Freeze 8. Other genetic data were used based on genotyping (rather than WGS) using an Illumina custom array, previously reported4. Genome-wide imputation was conducted using the multi-ethnic NHLBI Trans-Omics for Precision Medicine (TOPMed) freeze 8 reference panel20. Principal components (PCs) were previously computed using PC-Relate21, and the kinship matrix was computed using the genetic data. ‘Genetic analysis groups’ were constructed based on a combination of self-identified Hispanic/Latino backgrounds and genetic similarity, and are classified as Central American, Cuban, Dominican, Mexican, Puerto Rican, and South American4.
Ancestry enriched variants
We focused on a region of 6Mbp encompassing the APOE gene (~ 3Mbp from each side, mimicking the approach of Rajabli et al.9), chr19:42 Mb-48 Mb (GRCh38/hg38). Ancestry-specific frequencies of variants located in this region were calculated using GAFA11, a method we previously developed to estimate the frequencies of bi-allelic variants in admixed populations based on global proportions of genetic ancestries. Overall, the average ancestral global proportion of the three ancestries in the total dataset is 55% European, 30.5% Amerindian, and 14.5% African. Frequency estimation was based on n = 8933 HCHS/SOL individuals who consented to genetic data sharing with the broad scientific community. We defined Amerindian-enriched variants as those with both European and African minor allele frequency (MAF) < 0.01 and Amerindian frequency between 0.05 and 0.95. Using the same principle, we defined African-enriched variants. MAF is computed in a population, and it quantifies how likely it is for an individual from the population to have a specific genetic variant. A variant with a very low MAF (often MAF < 0.01 is considered rare) in a population is likely to be observed only in a few individuals in that population, and a variant with a high MAF (the largest possible value is 0.5) is likely to be observed in many individuals. Thus, an Amerindian (African) enriched variant is likely to be observed almost only in individuals who inherited the corresponding genomic region from an Amerindian (African) ancestor.
Bioinformatics
We performed bioinformatics analyses using publicly available databases and tools for the ancestry-enriched variants. Variant data were annotated using SnpEff (V.4.3). The SnpEff software takes as an input a genetic data file (variant calls on a VCF file) and it annotates each of the variants in the file using sequence ontology terms of predicted effects of the variants on known genes (e.g., codon deletion, exon duplication). It also provides impact prediction, with four categories: high, moderate, low, and modifier. Moderate impact means that the variant might change protein effectiveness, and high impact means that the variant is assumed to have high disruptive impact on the relevant protein. Low impact is annotated for variants that assumed to be harmless, and modifier impact is usually assigned for non-coding variants or variants affecting non-coding genes, where there is no evidence of impact. Variants with estimated moderate putative impact were selected for further analyses. We further annotated the variants by using additional tools: RegulomeDB22, GTEx Portal, GWAS catalog23, and Phegen24. Finally, we computed the MAF and minor allele counts (MAC) of each of the selected variants in the six Hispanic genetic analysis groups.
Statistical analysis
We provided descriptive statistics to characterize the demographic and cognitive outcome and APOE alleles distributions in the analytic dataset of n = 4,237 individuals. For each ancestry-enriched variant with a SnpEff predicted moderate impact, we tested the interaction associations between the variant and APOE-ε4 allele on MCI. Models included the variant, the APOE-ε4 allele (additive mode), and the interaction term of the variant with the APOE-ε4 allele. We used the complex survey design from the R ‘survey’ package25, with a “quasipoisson” family for binary traits. This method accounts for the stratification, clustering, and probability weighting in HCHS/SOL to allow correct generalizations to the HCHS/SOL target population. Models were adjusted for age, sex, education, center, first 5 PCs of genetic data, and genetic analysis group. The significance of the results was evaluated in two ways, to protect from potential high type 1 error due to the low proportion of APOE variant and the enriched variant alleles. First, through 5000 permutations, we performed multivariant Wald tests to jointly test the significance of the variant and variant-APOE-ε4 allele interaction. We also performed multivariant Wald tests to jointly test the association of the variant and variant-APOE-ε4 allele interaction and the APOE-ε4 allele. Second, we used mixed models and the BinomiRare test for low-count variants to test the association of the variant and variant-APOE-ε4 allele interaction26. Mixed models used correlation matrices to account for genetic relatedness (kinship), household, and block unit sharing as random effects, and were implemented, along with the BinomiRare, in the GENESIS R/Bioconductor package27, version 3.15.
Estimation of interaction associations with MCI in the ARIC study
We further evaluated the interaction associations between the African-enriched variant and APOE-ε4 in African Americans from the Atherosclerosis Risk in Communities (ARIC) study. ARIC is a longitudinal cohort study with genetic and cognitive measures28,29. The protocol for MCI/dementia diagnosis in ARIC has been previously described30 and is provided in Supplementary Note 1. Data from ARIC Visit 5 were used in this analysis. Further details are provided in Supplementary Note 1.
Similar to the model in SOL-INCA, the statistical model in ARIC included adjustment for age, sex, education, 5 PCs, and study-site, and tested the interaction associations between the variant and APOE-ε4 allele on a neurocognitive outcome. We also performed multivariant Wald tests as described above for SOL-INCA.
Results
Table 1 characterizes the demographic, health, and lifestyle characteristics of the SOL-INCA analytic dataset (n = 4237). Overall, around 52% of the participants are females, with a weighted mean age of 62 years at the SOL-INCA visit. MCI prevalence is ~ 11.3%. APOE-ε3 is the most frequent allele, with 0.83 allele frequency, while alleles 4 and 2 are relatively rare (frequencies of 0.12 and 0.049 respectively). Fifty-eight of the individuals with MCI (1.4% of the sample) were classified as having suspected severe impairment (MCI +).
Ancestry enriched variants
In the 6Mbp encompassing the APOE gene (~ 3Mbp from each side), chr19:42–48 Mb (GRCh38/hg38), we identified 260 Amerindian-enriched variants in the HCHS/SOL study population, with ≥ 5% frequency for Amerindian, and ≤ 1% frequency for African and European ancestries (Supplementary Table 1). Similarly, we identified 798 African-enriched variants in the HCHS/SOL study population (Supplementary Table 2). Using SnpEff (V.4.3) variant annotation, we selected 5 Amerindian- and 14 African-enriched variants, with a predicted moderate putative impact, for further interaction analyses with APOE-ε4 on MCI. Annotations of the variants and their estimated ancestral frequencies are presented in Table 2. Eighteen out of the total 19 selected enriched variants are missense variants. According to the GTExportal, 11 out of the 14 selected African-enriched variants have previously been associated with gene expressions in various tissues, i.e., they are expression quantitative trait loci (eQTLs) in these tissue, including brain cerebellum tissue.
Supplementary Table 3 provides the computed MAF and MAC of each of the selected variants across the genetic analysis groups corresponding to the six Hispanic backgrounds. As expected, Amerindian-enriched variants tended to be more common in groups with high Amerindian ancestry: Central and South American, and Mexican individuals, while African-enriched variants were more common in the Caribbean group, that have higher African ancestry: Dominican, Cuban, and Puerto Rican.
Interaction association between ancestry enriched variants and APOE alleles on MCI
Results of the enriched Amerindian or African variants’ interaction with APOE-ɛ4 on MCI analysis based on the 5,000 permutations multivariant Wald tests are reported in Table 3. No statistically significant interaction associations were identified. However, results of the enriched Amerindian or African variants’ interaction with APOE-ɛ4 on MCI analysis, the BinomiRare test identified one nominally significant interaction between an African enriched variant, rs8112679, and APOE-ɛ4 on MCI (p-value = 0.017) (Table 4).
Estimation of interaction associations with MCI in the ARIC study and meta-analysis
Supplementary Table 4 characterizes the demographic, cognitive outcome, and APOE alleles’ distributions in the ARIC African Americans analytic dataset. The African enriched variant’s interaction (rs8112679) with the APOE-ɛ4 on MCI was not significant in the ARIC African Americans analytic dataset (Supplementary Table 5).
A replication attempt of the interaction association between the African variant previously reported by Rajabli et al.9 and the APOE-Ɛ4 allele on MCI in the SOL-INCA analytic dataset did not present a significant result (Supplementary Table 6).
Discussion
In this study, we leveraged recently published data on ancestry-specific genetic variant frequencies in the Hispanic/Latino population11, to explore the interaction effects of African and Amerindian ancestry-enriched genetic variants with APOE-ɛ4 on MCI in the Hispanic/Latino US population. We found suggestive evidence for an interaction effect of an African-enriched variant, rs8112679, with APOE-ɛ4 on MCI, with the minor allele A having a protective effect on MCI. This result did not replicate in the ARIC African American analytic sample. Rs8112679 is a missense variant, located in exon 4 of the ZNF222 gene. ZNF222 gene is predicted to be involved in the regulation of transcription by RNA polymerase II. A previous study suggests the involvement of ZNF222 in late-onset Alzheimer’s disease31. Hopefully, future studies, particularly with African and African American individuals in which the variant is common and, ideally, a familial study with individuals both affected and unaffected by MCI or AD, would follow up on this variant and study its presence in affected individuals.
Our study is based on relatively small sample sizes therefore its statistical power is limited for the association of low-frequency and rare variants, all the more so for interaction analyses. Only large effect sizes could have been discovered. Our results suggest there are no large effect sizes of ancestry-enriched variants interacting with APOE-ɛ4 on MCI in the APOE region in the Hispanic/Latino population. Further analysis with larger sample sizes, and meta-analyses with additional studies, are needed to identify ancestry-enriched variants interacting with APOE-ɛ4 on MCI. It would also be interesting to increase the region in which variants are considered, potentially within other known AD genes or genome-wide. The major limitation is power, reduced by high multiple testing burden. Collaboration between multiple studies with diverse individuals with Amerindian and/or African ancestry will be critical. Another limitation of our study is that a subset of the MCI individuals was classified as MCI + , who fell into a gray zone between MCI and dementia. That is, their cognitive scores or functional abilities (i.e., IADLs) did not meet strict criteria for MCI or dementia. As SOL-INCA study population grows older, we will know whether the MCI and MCI + individuals convert to dementia. The cognitive aging trajectories of MCI is important and unanswered research question facing the field. It is possible that genetic determinants underlying mild and severe cognitive impairment are different. However, larger datasets are required to study this hypothesis, to assess trajectories of mild and severe cognitive impairment, and to identify trait-specific genetic basis.
Identification of variants interacting with APOE-ɛ4 may further delineate the role of APOE in the pathogenesis of MCI and AD and advance novel therapeutics. It may also lead to population-specific risk predictions and help reduce health disparities in the general population.
Data availability
HCHS/SOL genetic and phenotypic data can be obtained through the study's Data Coordinating Center using an approved data use agreement. Information is provided in https://sites.cscc.unc.edu/hchs/. HCHS/SOL genetic and phenotypic data can also be obtained from dbGaP under accession number phs000810.v1.p1. ARIC genetic and phenotypic data can be obtained through the study's Data Coordinating Center using an approved data use agreement. Information is provided in https://sites.cscc.unc.edu/aric/distribution-agreements.
References:
Tervo, S. et al. Incidence and risk factors for mild cognitive impairment: a population-based three-year follow-up study of cognitively healthy elderly subjects. Dement. Geriatr. Cogn. Disord. 17, 196–203 (2004).
Alzheimer Association. Alzheimer’s disease facts and figures. Alzheimer’s & Dementia 14, 367–429 (2018).
Dilworth-Anderson, P., Hendrie, H. C., Manly, J. J., Khachaturian, A. S. & Fazio, S. Diagnosis and assessment of Alzheimer’s disease in diverse populations. Alzheimers Dement. 4, 305–309 (2008).
Conomos, M. P. et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic community health study/study of Latinos. Am. J. Human Genet. 98, 165–184 (2016).
Granot-Hershkovitz, E. et al. APOE alleles’ association with neurocognitive function differ across Hispanic background groups. Alzheimers Dement. 16, e044169 (2020).
Hohman, T. J. et al. Global and local ancestry in African–Americans: implications for Alzheimer’s disease risk. Alzheimers Dement. 12, 233–243 (2016).
Rajabli, F. et al. Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 14, e1007791 (2018).
Blue, E. E., Horimoto, A., Mukherjee, S., Wijsman, E. M. & Thornton, T. A. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement. 15, 1524–1532 (2019).
Rajabli, F. et al. A locus at 19q1331 significantly reduces the ApoE ε4 risk for Alzheimer’s Disease in African Ancestry. PLoS Genet. 18, e1009977 (2022).
Le Guen, Y. et al. Association of rare APOE missense variants V236E and R251G With risk of Alzheimer disease. JAMA Neurol. 79, 652–663 (2022).
Granot-Hershkovitz, E. et al. AFA: ancestry-specific allele frequency estimation in admixed populations: the Hispanic community health study/study of Latinos. HGG Adv. 3, 100096 (2022).
Lavange, L. M. et al. Sample design and cohort selection in the Hispanic community health study/study of Latinos. Ann. Epidemiol. 20, 642–649 (2010).
Sorlie, P. D. et al. Design and implementation of the Hispanic community health study/study of Latinos. Ann. Epidemiol. 20, 629–641 (2010).
Albert, M. S. et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 270–279 (2011).
González, H. M. et al. A research framework for cognitive aging and Alzheimer’s disease among diverse US Latinos: design and implementation of the Hispanic Community Health Study/Study of Latinos-Investigation of Neurocognitive Aging (SOL-INCA). Alzheimers Dement. 15, 1624–1632 (2019).
González, H. M. et al. Prevalence and correlates of mild cognitive impairment among diverse hispanics/latinos: Study of Latinos-Investigation of Neurocognitive Aging results. Alzheimers Dement. 15, 1507–1515 (2019).
Tomaszewski Farias, S. et al. The measurement of everyday cognition: development and validation of a short form of the Everyday Cognition scales. Alzheimers Dement. 7, 593–601 (2011).
González, H. M. et al. Neurocognitive function among middle-aged and older Hispanic/Latinos: results from the Hispanic Community Health Study/Study of Latinos. Arch. Clin. Neuropsychol. 30, 68–77 (2015).
González, H. M. et al. Apolipoprotein E genotypes among diverse middle-aged and older Latinos: Study of Latinos-Investigation of Neurocognitive Aging results (HCHS/SOL). Sci. Rep. 8, 17578 (2018).
Kowalski, M. H. et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 15, e1008500 (2019).
Conomos, M. P., Reiner, A. P., Weir, B. S. & Thornton, T. A. Model-free estimation of recent genetic relatedness. Am. J. Human Genet. 98, 127–148 (2016).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Ramos, E. M. et al. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Human Genet. 22, 144–147 (2014).
Lumley, T. & Scott, A. Tests for regression models fitted to survey data. Aust. N. Z. J. Stat. 56, 1–14 (2014).
Sofer, T. et al. BinomiRare: a robust test for association of a rare genetic variant with a binary outcome for mixed models and any case-control proportion. Human Genet. Genom. Adv. 2(3), 100040. https://doi.org/10.1016/j.xhgg.2021.100040 (2021).
Gogarten, S. M. et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics 35(24), 5346–5348. https://doi.org/10.1093/bioinformatics/btz567 (2019).
The ARIC investigators. The atherosclerosis risk in communities (ARIC) study: design and objectives. Am. J. Epidemiol. 129, 687–702 (1989).
Wright, J. D. et al. The ARIC (atherosclerosis risk in communities) study: JACC focus seminar 3/8. J. Am. Coll. Cardiol. 77, 2939–2959 (2021).
Knopman, D. S. et al. Mild cognitive impairment and dementia prevalence: the atherosclerosis risk in communities neurocognitive study (ARIC-NCS). Alzheimers Dement. (Amst) 2, 1–11 (2016).
Rao, S. et al. An APOE-independent cis-eSNP on chromosome 19q13.32 influences tau levels and late-onset Alzheimer’s disease risk. Neurobiol. Aging 66, e171–e178 (2018).
Acknowledgements
The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website—http://www.cscc.unc.edu/hchs/. The Hispanic Community Health Study/Study of Latinos is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I / N01-HC-65233), University of Miami (HHSN268201300004I / N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I / N01-HC-65235), University of Illinois at Chicago (HHSN268201300003I / N01- HC-65236 Northwestern Univ), and San Diego State University (HHSN268201300005I / N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. This work was supported by the National Institute on Aging (R01AG048642, RF1AG054548, RF1AG061022, R21AG070644, and R21AG056952). Dr. González also receives additional support from P30AG062429 and P30AG059299. The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Neurocognitive data is collected by U01 2U01HL096812, 2U01HL096814, 2U01HL096899, 2U01HL096902, 2U01HL096917 from the NIH (NHLBI, NINDS, NIA and NIDCD). The authors thank the staff and participants of the ARIC study for their important contributions.
Author information
Authors and Affiliations
Contributions
E.G.H. and B.S. performed genetic association analysis in HCHS/SOL. Y.Y. and R.X. performed genetic association analyses in ARIC. E.G.H. designed the analysis and drafted the manuscript. T.S. supervised the analysis. M.D., M.A., R.K., C.D., H.M.G. worked on HCHS/SOL and SOL-INCA data design and collection. J.C. worked on HCHS/SOL data curation. R.B.L. and M.F. worked on ARIC data design and collection. E.G.H, Y.Y., R.X., B.S., W.T, P.M.V., R.B.L., M.D., M.A., J.C., R.K., M.F., C.D., H.M.G., and T.S. critically reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Granot-Hershkovitz, E., Xia, R., Yang, Y. et al. Interaction analysis of ancestry-enriched variants with APOE-ɛ4 on MCI in the Study of Latinos-Investigation of Neurocognitive Aging. Sci Rep 13, 5114 (2023). https://doi.org/10.1038/s41598-023-32028-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32028-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.