Abstract
Respiratory diseases are leading causes of mortality and morbidity worldwide. Pulmonary imaging is an essential component of the diagnosis, treatment planning, monitoring, and treatment assessment of respiratory diseases. Insights into numerous pulmonary pathologies can be gleaned from functional lung MRI techniques. These include hyperpolarized gas ventilation MRI, which enables visualization and quantification of regional lung ventilation with high spatial resolution. Segmentation of the ventilated lung is required to calculate clinically relevant biomarkers. Recent research in deep learning (DL) has shown promising results for numerous segmentation problems. Here, we evaluate several 3D convolutional neural networks to segment ventilated lung regions on hyperpolarized gas MRI scans. The dataset consists of 759 helium-3 (3He) or xenon-129 (129Xe) volumetric scans and corresponding expert segmentations from 341 healthy subjects and patients with a wide range of pathologies. We evaluated segmentation performance for several DL experimental methods via overlap, distance and error metrics and compared them to conventional segmentation methods, namely, spatial fuzzy c-means (SFCM) and K-means clustering. We observed that training on combined 3He and 129Xe MRI scans using a 3D nn-UNet outperformed other DL methods, achieving a mean ± SD Dice coefficient of 0.963 ± 0.018, average boundary Hausdorff distance of 1.505 ± 0.969 mm, Hausdorff 95th percentile of 5.754 ± 6.621 mm and relative error of 0.075 ± 0.039. Moreover, limited differences in performance were observed between 129Xe and 3He scans in the testing set. Combined training on 129Xe and 3He yielded statistically significant improvements over the conventional methods (p < 0.0001). In addition, we observed very strong correlation and agreement between DL and expert segmentations, with Pearson correlation of 0.99 (p < 0.0001) and Bland–Altman bias of − 0.8%. The DL approach evaluated provides accurate, robust and rapid segmentations of ventilated lung regions and successfully excludes non-lung regions such as the airways and artefacts. This approach is expected to eliminate the need for, or significantly reduce, subsequent time-consuming manual editing.
Similar content being viewed by others
Introduction
Respiratory diseases are leading causes of mortality and morbidity worldwide with 339 million experiencing asthma, 65 million people with chronic obstructive pulmonary disease (COPD)1,2 and 1.8 million new lung cancer cases diagnosed every year3. Pulmonary imaging, using various modalities, is an essential part of the diagnosis, treatment planning, monitoring, and treatment assessment of respiratory diseases. The acquisition, processing, and interpretation of pulmonary images are critical components of patient management and are essential in reducing mortality and morbidity.
Currently, computed tomography (CT) is the clinical gold standard for pulmonary imaging due to its exceptional spatial and temporal resolution, and its ubiquitous availability. CT is a structural imaging modality that provides exquisite detail of morphological changes in the lung parenchyma but employs ionizing radiation. Although proton magnetic resonance imaging (1H MRI) has historically been susceptible to the low proton density in lungs, recent advances in pulse sequences and hardware with ultra-short and zero echo times have enabled 1H MRI to compete with CT with the added benefit of no ionizing radiation4,5. However, whilst structural imaging modalities facilitate the assessment of changes in lung tissue density, they do not directly provide an accurate picture of regional lung function.
Although nuclear imaging modalities such as single-photon emission computed tomography (SPECT) can provide regional lung function information6, they require harmful ionizing radiation, reducing the ability to conduct regular scans during clinical care. This is particularly important when imaging children, as developing tissue is more sensitive to ionizing radiation. Moreover, SPECT is limited by poor temporal and spatial resolution and images acquired using 99mTc-diethylenetriamine pentaacetate (DTPA) aerosols, one of the most commonly used radiotracers for ventilation imaging with SPECT, are subject to clumping artefacts6,7. In contrast, unparalleled insights into respiratory diseases can be gleaned from non-ionizing functional lung MRI modalities, such as dynamic contrast-enhanced lung perfusion MRI and hyperpolarized gas ventilation MRI. Hyperpolarized gas MRI provides visualization and quantification of regional lung ventilation with high spatial resolution within a single breath8. Quantitative biomarkers derived from this modality, including the ventilated defect percentage (VDP) and coefficient of variation, provide further insights into regional ventilation9,10,11. To facilitate the computation of such biomarkers, segmentation of ventilated regions of the lungs is required12.
Previous approaches for hyperpolarized gas MRI ventilation segmentation employed classical image processing and machine learning approaches, such as hierarchical K-means13 and spatial fuzzy c-means (SFCM) clustering14. However, as these methods rely on voxel intensities and thresholding, they only provide semi-automatic segmentations; as such, they are prone to generate errors in regions where voxel intensities are similar to those of the ventilated lung region (e.g., airways and artefacts). Consequently, they frequently require significant time to manually correct.
Deep learning (DL), which utilizes artificial neural networks with multiple hidden layers, has shown tremendous promise in medical image segmentation applications15. Although DL was initially theorized over half a century ago, the field only received widespread acclaim in 2012 when AlexNet, a form of an artificial neural network referred to as a convolutional neural network (CNN), triumphed in the ImageNet Large Scale Visual Recognition Challenge16. Subsequently, CNNs, and DL more generally, have become mainstream in the medical image segmentation field. UNet and VNet CNNs have demonstrated their profound impact in numerous medical image segmentation problems17,18. Adoption has been enhanced through transfer learning to cope with limited datasets common in the medical imaging field19. In a recent review of DL-based lung image analysis studies, Astley et al. identified a significant gap in DL-based lung MRI segmentation studies (n = 7) with only one published conference proceeding20 and one journal article21 evaluating DL for hyperpolarized gas MRI segmentation. Tustison et al. used a 2D UNet for hyperpolarized gas MRI segmentation on a dataset of 113 images, developing a novel template-based method to augment the limited lung imaging data alongside pre-processing techniques, including N4 bias correction and adaptive denoising. A mean ± SD DSC between DL and manual segmentations of 0.94 ± 0.03 was achieved21. However, the application of DL on a more extensive dataset with a broader range of pathologies is required prior to clinical adoption.
In this work we conducted extensive parameterization experiments to determine the best-performing 3D CNN architecture, loss function and pre-processing techniques for hyperpolarized gas MRI segmentation. We further evaluated five DL methods using the best performing configuration to accurately, robustly and rapidly segment ventilated lungs on hyperpolarized gas MRI scans. Using a diverse testing set, with both helium-3 (3He) and xenon-129 (129Xe) noble gas scans and corresponding expert segmentations, we evaluated and compared performance using a range of evaluation metrics. We also investigated the effect of the noble gas on DL performance. Furthermore, we compared the best performing DL method to conventional approaches. Finally, ventilated lung volume correlation and agreement were assessed for the best-performing DL method compared to expert-derived volumes.
Materials and methods
Hyperpolarized gas MRI acquisition
All subjects underwent 3D volumetric 3He or 129Xe hyperpolarized gas MRI with full lung coverage at 1.5 T on a HDx scanner (GE Healthcare, Milwaukee, WI) using 3D steady-state free precession (SSFP) sequences as previously described22,23,24. Flexible quadrature radiofrequency coils were employed for transmission and reception of MR signals at the Larmor frequencies of 3He and 129Xe. In-plane (x–y) resolution of scans for both gases was 4 × 4mm2. 129Xe scans ranged from 16 to 34 slices with a mean of 23 slices and slice thickness of 10 mm. 3He scans ranged from 34 to 56 slices with a mean of 45 slices and slice thickness of 5 mm.
Dataset
The imaging dataset used in this study was pooled retrospectively from several research studies and clinical studies of patients referred for hyperpolarized gas MRI scans. Data use was approved by the Institutional Review Boards at the University of Sheffield and the National Research Ethics Committee. All data was anonymized and all investigations were conducted in accordance with the relevant guidelines and regulations.
The dataset consisted of 759 volumetric hyperpolarized gas MRI scans (23,265 2D slices), with either 3He (264 scans, 11,880 slices) or 129Xe (495 scans, 11,385 slices), from 341 subjects. The slices were distributed approximately 50:50 between 3He and 129Xe. The dataset contained healthy subjects and patients with various pulmonary pathologies: asthma, COPD, asthma/COPD overlap, bronchiectasis, interstitial lung disease (ILD), idiopathic pulmonary fibrosis (IPF), lung cancer, cystic fibrosis (CF), children born prematurely, and patients investigated for possible airway disease. Demographic and clinical data for these subjects are summarized in Table 1.
Each of the 759 scans in the dataset has a corresponding, manually-edited expert segmentation, representing the ventilated region of the lungs. These scans and segmentations were collected from numerous retrospective studies; consequently, the segmentations were generated using several semi-automated methods14,25 and edited by multiple expert observers. Quality control was conducted by an experienced imaging scientist who identified potential errors and manually corrected them to ensure segmentation accuracy; the airways were removed down to the third generation, and it was ensured that no voxels were outside of the lung parenchymal region defined by a structural 1H MRI scan, thereby removing background noise.
Convolutional neural network
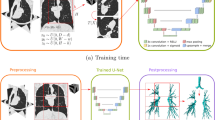
Parameterization experiments were conducted comparing network architectures, loss functions and pre-processing techniques; results of these experiments are provided in the Supplementary Information. We used the nn-UNet fully convolutional neural network which processes 3D scans using volumetric convolutions. The network is trained end-to-end using hyperpolarized gas MRI volumetric scans. We use a 3D implementation of the UNet which has been modified to reduce memory constraints, allowing 30 feature channels26. Convolution operations vary in size from 3 × 3 × 3 to 1 × 1 × 1 depending on the layer of the network. The network also makes use of instance normalization. An isotropic spatial window size was used of [96, 96, 96] with a batch size of 2. A high-level visual representation of the 3D nn-UNet, specific to the spatial window sizes used, is shown in Fig. 1.
Parameters
The network utilizes a non-linear PReLU activation function27 and is optimized using a binary cross-entropy loss function. ADAM optimization was used to train the CNN28 and instance normalization was conducted for each pass. The spatial window size was [96, 96, 96] with a batch size of 2. A learning rate of 1 × 10−5 was used for initial training and 0.5 × 10−5 for subsequent fine-tuning methods.
Pre-processing
Each hyperpolarized gas MRI scan was pre-processed using spatially adaptive denoising, designed to consider both Rician noise and spatially varying patterns of noise. Denoising was implemented with ANTs 2.1.0 using the DenoiseImage function across three dimensions. Standard parameters were used29.
Data augmentation
Constrained random rotation and scaling was used for data augmentation. Rotation with limits − 10° to 10° and scaling of − 10 to 10%, where a random rotation or scaling were applied at an interval within those limits, were used. A different random value was computed for each rotation axis and scaling factor.
Data split
The dataset was randomly split into training and testing sets with 75% and 25% of the data respectively, in terms of the number of subjects. The training set, therefore, contained 237 3He scans (10,902 slices) and 436 129Xe scans (10,028 slices) from a total of 255 subjects. 86 scans, each from a different subject, were selected for the testing set (3He: 27 scans (1242 slices); 129Xe: 59 scans (1357 slices)). Repeat or longitudinal scans from multiple visits for the same patient were contained in the training set; however, no subject was present in both the training and testing sets, with the testing set containing only one scan from each patient. This was ensured by randomly selecting only one scan from each subject in the testing set and discarding the remaining scans; these scans are not included in Table 1. The range of diseases in the testing set was representative of the dataset as a whole. In addition, it was specified that there would be no overlap between the newly defined testing set and the previous testing set used for parameterization experiments, described in the Supplementary Information, in terms of either patient or scan.
Computation
All networks were trained using the medical imaging DL framework NiftyNet 0.6.030 built on top of TensorFlow 1.1431. Training and inference were performed on an NVIDIA Tesla V100 GPU with 16 GB of RAM.
DL experimental methods
Five DL experimental methods were performed to train the network:
-
(1)
The model was trained on 237 3He scans for 30,000 iterations.
-
(2)
The model was trained on 436 129Xe scans for 30,000 iterations.
-
(3)
The model was trained on 237 3He scans for 20000 iterations; these weights were used to initialize a model trained on 436 129Xe scans for 10000 iterations.
-
(4)
The model was trained on 436 129Xe scans for 20000 iterations; these weights were used to initialize a model trained on 237 3He scans for 10000 iterations.
-
(5)
The model was trained on 436 129Xe and 237 3He scans for 30,000 iterations.
The five experimental methods were applied to the data split defined above using the same testing set for each method, facilitating comparison between the five methods to identify the best performing training method across multiple metrics.
Comparison to conventional methods
For further benchmarking, the best-performing DL method was compared against two other conventional machine learning methods, namely, SFCM and K-means clustering. The methods used are described as follows:
-
(1)
The k-means clustering algorithm used here was previously modified for hyperpolarized gas MRI segmentation32. This method attempts to find k data points, given the integer k, in an n-dimensional space (Rn) given m data points. These k data points are known as centres/centroids and the aim is to minimize the distance from each data point (m) to its centre/centroid33. The previously developed method32 attempts to delineate the image data into a number of clusters that can best represent a radiologist’s analysis of the ventilation image with clusters defined from defects to hyperintense signal. The first stage of this method requires image normalization into the range of 0–255, following which the cluster initial centers are set at 25% intervals between these values. A two-stage clustering process was applied with four clusters in the first stage, the lowest of which contains both signal void and hypointense signal. In the second stage, the clustering was reapplied to the lowest cluster from the first stage to define background, ventilation defect and hypointense signal regions.
-
(2)
The SFCM method used in this work has been reported previously14; images are initially filtered to remove noise and maintain edges using a bilateral filter34. The standard FCM algorithm assigns N pixels to C clusters via Fuzzy memberships. The key assumption of the Spatial Fuzzy C-means is that pixels spatially close will have high correlation and hence have similarly high membership to the same cluster. This spatial information will modify the membership value only if, for example, the pixel is noisy and would have been incorrectly classified. The SFCM method makes use of nearby pixels during the iteration process by taking into account the membership of voxels within a predefined window (5×5 in this work) and will weight the central pixel depending on the provided weighting variables35. The optimal number of clusters was manually selected by the observer.
Evaluation Metrics
The testing set results for each of the five DL experimental methods and two conventional methods were evaluated using several metrics. The DSC was used to assess overlap between the ground truth (GT) and predicted (PR) segmentations36 and is defined as:
Two distance metrics, average boundary Hausdorff distance (Avg HD) and 95th percentile Hausdorff distance (HD95) were used37 and are defined as the following:
where h(PR, GT) represents the directed Hausdorff distance between the sets of PR and GT voxels at the boundary, pr represents an individual voxel in the set PR and gt represents an individual boundary voxel in GT. h(PR, GT) is defined as:
\(\left\| {PR - GT} \right\|\) is the Euclidean distance between PR and GT. From this, HD95 is calculated as the 95th percentile of Eq. (2) and is frequently used in the image segmentation literature to remove the impact of outlier voxels. The Avg HD is defined similarly as:
where d(PR, GT) represents the directed average Hausdorff distance given by:
where N is the set of paired voxels in (GT, PR). The Avg HD reduces sensitivity to outliers and is regarded as a stable metric for segmentation evaluation38.
Furthermore, a relative error metric (XOR) was used to evaluate segmentation errors39 as follows:
where PR’ and GT’ are the complements of PR and GT, respectively. The metric was used specifically because it is expected to correlate with the manual editing time required to correct the segmentation outcome.
Statistical analysis
Data were tested for normality using Shapiro–Wilk tests; when normality was not satisfied, non-parametric tests were conducted. One-way repeated-measure ANOVA or Friedman tests were conducted as appropriate with Bonferroni correction for post-hoc multiple comparisons to assess statistical significances of differences between experimental DL-based methods. Independent t-tests or Mann–Whitney U tests were used to compare differences between 3He and 129Xe segmentations in the testing set, assessing the effect of the noble gas. The best performing DL method was compared to other conventional segmentation methods using one-way repeated-measure ANOVA or Friedman tests with Bonferroni correction for post-hoc multiple comparisons. Pearson or Spearman correlations and Bland–Altman analysis were conducted to compare volumes of DL-generated and expert segmentations. Statistical analysis was performed using Prism 8.4 (GraphPad, San Diego, CA) and SPSS Statistics 26.0 (IBM Corporation, Armonk, NY).
Results
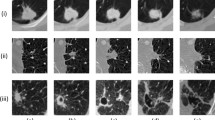
Segmentations for each of the five DL methods were generated for 86 testing set scans. Figure 2 shows examples of segmentation quality for a healthy subject and patients with four different pathologies across the five DL experimental methods using 3He or 129Xe. The original scans and expert segmentations are included to facilitate comparison. It can be observed that, in general, there are negligible voxels outside the lung parenchyma classed as ventilated and that the CNNs accurately excluded ventilation defects, as shown in the examples of the asthma and lung cancer patients. Case 4, of a healthy subject, represents an interesting case due to the presence of a zipper artefact caused by electronic noise in the hardware; it can be observed that some models are able to accurately exclude this artefact, whilst others remain unable to distinguish between the zipper artefact and ventilated lung voxels. Table 2 summarizes segmentation performance for the five DL experimental methods. The Combined 3He and 129Xe method generated the best segmentations using all four metrics.
Figure 3 shows distributions of all four metrics for each DL method. The assumption of normality for each metric was not satisfied for all DL methods, as assessed by Shapiro–Wilk’s tests (p < 0.05). As such, Friedman tests were run, determining that there were differences between DL methods for each metric. Post-hoc pairwise comparisons were performed for each metric with Bonferroni correction for multiple comparisons. The combined 3He and 129Xe method yielded statistically significant improvements over all DL methods using the DSC, XOR and HD95 metrics (p < 0.05). However, using the Avg HD metrics, the combined 3He and 129Xe method significantly outperformed all but one DL method.
Comparison of segmentation performance on 86 testing scans for five DL experimental methods using the DSC, Avg HD, HD95 and XOR metrics (left to right). P-values are displayed for Friedman tests with Bonferroni correction for multiple comparisons, comparing the combined 3He and 129Xe DL method to the other DL methods. Mean and standard deviation values are marked by a bold line and whiskers, respectively.
Figure 4 shows the segmentation performance for the testing set stratified by noble gas (129Xe or 3He) using the DSC and Avg HD metrics. The majority of models show no significant difference between 129Xe and 3He for both metrics. Only two methods, namely, the ‘Train on 3He’ and ‘Train on 129Xe, fine-tune on 3He’ methods, showed a significant difference between noble gases across both metrics.
Based on the results of the five experimental methods, the combined 3He and 129Xe DL model was identified as the most accurate DL ventilated lung segmentation method due to statistically significant improvements over all other methods using the DSC, HD95 and XOR metrics. Consequently, we tested the combined 3He and 129Xe DL model on 31 2D spoiled gradient-echo 3He hyperpolarized gas MRI ventilation scans which differ in MRI sequence and acquisition parameters (see Supplemental Results). The results indicated that the model generalized to scans acquired with a different MRI sequence and produced high quality segmentations invariant of the sequence used.
Furthermore, ventilated volume was assessed for the combined 3He and 129Xe method. The assumption of normality of was satisfied for DL and expert ventilated volume, as assessed by Shapiro–Wilk’s tests (p > 0.05). Pearson correlation and Bland–Altman analysis are shown in Fig. 5 for the combined 129Xe and 3He model; the DL segmentation volume is highly correlated (r = 0.99) with the expert segmentation volume and exhibits minimal bias (− 0.8%).
Figure 6 shows qualitative and quantitative performance for the DL combined 3He and 129Xe training method with two conventional segmentation methods, namely K-means clustering and SFCM across three cases. The assumption of normality for the DSC metric was not satisfied for conventional and DL approaches, as assessed by Shapiro–Wilk’s tests. Post hoc Friedman’s tests were performed with Bonferroni correction for multiple comparisons (X2(3), p < 0.0001). The DL segmentation method exhibited significant improvements over conventional methods (p < 0.0001), accurately excluding low-level noise and artefacts (e.g. Case 2) as well as non-lung regions such as the trachea and bronchi.
Comparison of performance on testing scans between the combined 129Xe and 3He DL method and conventional segmentation methods (SFCM and K-means) with P-values for Friedman tests with Bonferroni correction for multiple comparisons. Mean and standard deviation values are marked by a bold line and whiskers, respectively. Individual DSC and Avg HD values for each method are displayed for three cases.
Discussion
The DL segmentation methods yielded highly accurate segmentations across a range of evaluation metrics on the dataset used. To the best of the authors’ knowledge, the hyperpolarized gas MRI dataset used here is the largest to date for ventilated lung segmentation, comprising 759 scans from patients with a wide range of lung pathologies. This is advantageous for preserving generalizability as it enables algorithms to learn features present in a range of diseases independent of the noble gas. Compared with 129Xe MRI, 3He MRI has an intrinsically stronger MRI signal due to the difference in gyromagnetic ratios between the two nuclei. Generally, lung ventilation information of similar diagnostic quality has been obtained with the two nuclei; despite this, there are known differences in lung diffusivity as well as differences in spatial resolution between the nuclei40,41. This is particularly important for deep learning applications as the resolutions of our 3He and 129Xe MRI scans differ greatly in the z-direction whereby 3He and 129Xe MRI scans have a slice thickness and an inter-slice distance of ~ 5 mm and ~ 10 mm, respectively. Therefore, it remains important to understand the performance of deep learning segmentation applications across the two nuclei.
The combined 3He and 129Xe DL method showed statistically significant improvements over all other methods using the DSC, HD95 and XOR metrics; however, using the Avg HD metric, no significant difference between the combined 3He and 129Xe method and 129Xe only method was observed, perhaps attributable to an outlier case. Some statistically significant differences were observed in performance when comparing 3He and 129Xe testing set scans; however, the combined 3He and 129Xe method exhibited identical performance independent of the noble gas used. This indicates that, for a given 3He or 129Xe scan, the combined 3He and 129Xe method is unlikely to be biased towards a specific noble gas. Due to the current paucity and unpredictable supplies of 3He worldwide, the field, in general, has transitioned towards the use of 129Xe as the predominant noble gas for hyperpolarized gas MR ventilation imaging. As this trend continues, it may be pertinent in future work to assess the impact of training and testing solely on 129Xe scans. In addition, external testing, detailed in the Supplemental Results, indicated the proposed model’s ability to generalize across MRI sequence and acquisition parameters not seen in the training set, further reinforcing that the model is using functional features from hyperpolarized gas MRI to produce accurate segmentations.
The CNN produced more accurate segmentations than the two conventional approaches for all evaluation metrics. In particular, the CNN was able to deal with images containing background noise and artefacts, as well as successfully excluding ventilation defects and airways. In comparison, the SFCM method was unable to distinguish airways or artefacts and segmented these areas erroneously. As such, it is highly probable that the CNN eliminates or dramatically reduces the manual-editing time required after automatic segmentation. The K-means clustering algorithm exhibited poorer than expected performance, possibly attributable to the lack of an available proton MRI. This represents a benefit of the CNN-based method as only the hyperpolarized gas MRI scan is required as an input. Previous work in the literature that aimed to employ DL for hyperpolarized gas MRI segmentation used a 2D UNet and achieved a mean DSC of 0.9421. In comparison, our combined 3He and 129Xe method trained via a 3D nn-UNet yielded a mean DSC value of 0.96. The 3D CNN allows the model to treat the segmentation as a 3D volume and learns features present across multiple slices e.g. ventilation defects. Several pre-processing techniques have previously been used in the literature for lung image segmentation42. The work of Tustison, et al.21 utilizes a novel template-based data augmentation strategy with N4 bias correction and denoising, which are computationally expensive and time-consuming; however, the impact of such techniques is not assessed in their work. In this study, we observed that N4 bias correction provided no significant benefit, while denoising yielded significant improvements.
All DL methods were trained and tested using a single GPU. Training required approximately nine days, while inference took 27 s per 129Xe scan and 35 s per 3He scan, corresponding to approximately one second per slice for both gases. Compared with conventional methods, such as SFCM, the time taken to generate automatic segmentations is significantly reduced from approximately 5 min to around 30 s, indicating the time-saving benefits of DL-based methods. Moreover, accurate automatic segmentation of hyperpolarized gas MRI ventilation scans through CNN-based approaches will eliminate or reduce manual editing time, thus improving clinical throughput. To further improve clinical translation of DL-based techniques, we have provided the trained DL model along with necessary files and Supplementary Instructions, enabling members of the pulmonary imaging community to apply the trained model in their own research.
The specific dataset used is unique within the context of pulmonary imaging due to the presence of numerous features such as different noble gases, longitudinal scans, repeat scans and pre- and post-treatment scans. The variation in the number of repeat or longitudinal scans and slice thicknesses between 3D 3He and 129Xe scans impeded us from achieving a training and testing set split equally between both gases; notwithstanding, the number of 2D slices were approximately equal between gases. Although multiple scans from the same patient were included in the training set to increase dataset numbers, to enhance the robustness of the evaluation, no scan of the same patient was present both in the training and testing sets.
This study also represents the first large-scale investigation of architectures, loss functions and pre-processing techniques within the field of lung MRI. Selecting a subset of the data allowed us to perform parameterization experiments to determine the ideal choice of network architecture, loss function and pre-processing technique, without creating optimization biases in subsequent experiments (see Supplementary Information). The conclusions of the parameterization experiments were partially limited due to multiple factors; the same exact parameters cannot be used for each network due to the spatial imaging constraints of the specific network, such as requiring isotropic resolutions or the varying memory requirements of each architecture. This means that the windowing, batch size and bordering varies between architectures and can, therefore, make comparisons potentially difficult. However, where possible, we aimed to maintain consistent parameters across all networks tested. Further investigation may aim to optimize other hyperparameters that could be deemed equally important as the experiments conducted related to architecture, loss function and pre-processing; these may include the choice of activation function or optimization algorithm. Furthermore, parameterization results will vary based on the specific datasets used and, consequently, limit conclusions to the particular data used in these experiments.
Currently, segmentations edited by expert observers are the gold-standard for training supervised DL algorithms. Studies have shown that manual segmentations are susceptible to inter-observer variability43. Numerous research projects have employed techniques to create generalizable DL models across multiple institutions and observers44. A limitation of our study is the presence of only one expert segmentation per scan, which precludes the ability to evaluate intra- and inter-observer variability. However, the wide range of expert observers used to generate and manually edit the expert segmentations led to significant variability in the training and testing sets. Hence, the CNN can learn a robust segmentation method invariant to the specific semi-automated method used to generate the ground truth or the expert observer who manually corrected it. In future work, multiple expert segmentations may be used to train the algorithm and allow the evaluation of inter-observer variability.
For the evaluation of certain clinically relevant metrics such as VDP9, the whole-lung cavity volume is required in addition to ventilated lung volumes, most commonly computed from a whole-lung segmentation generated from a structural proton MRI scan. In this work, we showed that ventilated lung volumes derived from CNN-generated segmentations have a significant Pearson correlation of 0.99 and a minimal Bland–Altman bias of − 0.8% with expert volumes. However, evaluation of DL-based methods using not only ventilated lung volume, but also VDP, would further the extensive validation required for clinical adoption.
Conclusion
In conclusion, we evaluated a 3D fully connected CNN using the nn-UNet architecture that is capable of producing accurate, robust and rapid hyperpolarized gas MRI segmentations on a large, diverse dataset. We compared five experimental DL methods and observed that combining 3He and 129Xe scans during training produces significantly improved segmentations. Compared with expert segmentations, this CNN-based method also showed a strong Pearson correlation and limited bias using Bland–Altman analysis. In addition, the CNN-based segmentation method significantly outperformed two conventional segmentation methods commonly used in the literature.
Data availability
The imaging datasets generated and/or analysed during the current study are not publicly available as they were generated as part of an industrial collaborative study that is still underway. Requests for data should be addressed to J.M.W.
References
Vos, T. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259. https://doi.org/10.1016/S0140-6736(17)32154-2 (2017).
GBD15, Wang, H. D., Naghavi, M. & Allen, C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544. https://doi.org/10.1016/S0140-6736(16)31012-1 (2016).
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. https://doi.org/10.3322/caac.21262 (2015).
Togao, O., Tsuji, R., Ohno, Y., Dimitrov, I. & Takahashi, M. Ultrashort echo time (UTE) MRI of the lung: Assessment of tissue density in the lung parenchyma. Magn. Reson. Med. 64, 1491–1498. https://doi.org/10.1002/mrm.22521 (2010).
Bae, K. et al. Comparison of lung imaging using three-dimensional ultrashort echo time and zero echo time sequences: Preliminary study. Eur. Radiol. 29, 2253–2262. https://doi.org/10.1007/s00330-018-5889-x (2019).
Petersson, J., Sánchez-Crespo, A., Larsson, S. A. & Mure, M. Physiological imaging of the lung: Single-photon-emission computed tomography (SPECT). J. Appl. Physiol. 102, 468–476. https://doi.org/10.1152/japplphysiol.00732.2006 (2007).
Yuan, S. T. et al. Semiquantification and classification of local pulmonary function by V/Q single photon emission computed tomography in patients with non-small cell lung cancer: Potential indication for radiotherapy planning. J. Thorac. Oncol. 6, 71–78. https://doi.org/10.1097/jto.0b013e3181f77b40 (2011).
Fain, S. B. et al. Functional lung imaging using hyperpolarized gas MRI. J. Magn. Reson. Imaging 25, 910–923. https://doi.org/10.1002/jmri.20876 (2007).
Woodhouse, N. et al. Combined helium-3/proton magnetic resonance imaging measurement of ventilated lung volumes in smokers compared to never-smokers. J. Magn. Reson. Imaging 21, 365–369. https://doi.org/10.1002/jmri.20290 (2005).
Tzeng, Y. S., Lutchen, K. & Albert, M. The difference in ventilation heterogeneity between asthmatic and healthy subjects quantified using hyperpolarized 3He MRI. J. Appl. Physiol. 1985(106), 813–822. https://doi.org/10.1152/japplphysiol.01133.2007 (2009).
Hughes, P. J. C. et al. Assessment of the influence of lung inflation state on the quantitative parameters derived from hyperpolarized gas lung ventilation MRI in healthy volunteers. J. Appl. Physiol. 126, 183–192. https://doi.org/10.1152/japplphysiol.00464.2018 (2019).
Tustison, N. J. et al. Ventilation-based segmentation of the lungs using hyperpolarized 3He MRI. J. Magn. Reson. Imaging 34, 831–841. https://doi.org/10.1002/jmri.22738 (2011).
Kirby, M. et al. Hyperpolarized He-3 magnetic resonance functional imaging semiautomated segmentation. Acad. Radiol. 19, 141–152. https://doi.org/10.1016/j.acra.2011.10.007 (2011).
Hughes, P. J. C. et al. Spatial fuzzy c-means thresholding for semiautomated calculation of percentage lung ventilated volume from hyperpolarized gas and (1) H MRI. J. Magn. Reson. Imaging 47, 640–646. https://doi.org/10.1002/jmri.25804 (2018).
Hesamian, M. H., Jia, W., He, X. & Kennedy, P. Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit. Imaging 32, 582–596. https://doi.org/10.1007/s10278-019-00227-x (2019).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. in 26th Annual Conference on Neural Information Processing Systems. 1097–1105 (2012).
Bakator, M. & Radosav, D. Deep learning and medical diagnosis: A review of literature. Multimodal Technol. Interact. 2, 47. https://doi.org/10.3390/mti2030047 (2018).
Lundervold, A. S. & Lundervold, A. Vol. 29 102–127 (Elsevier GmbH, 2019).
Tajbakhsh, N. et al. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning?. IEEE Trans Med Imaging 35, 1299–1312. https://doi.org/10.1109/TMI.2016.2535302 (2016).
Astley, J. R. et al. in Thoracic Image Analysis. (eds Jens Petersen et al.) 24–35 (Springer).
Tustison, N. J. et al. Convolutional neural networks with template-based data augmentation for functional lung image quantification. Acad. Radiol. 26, 412–423. https://doi.org/10.1016/j.acra.2018.08.003 (2019).
Horn, F. C. et al. Lung ventilation volumetry with same-breath acquisition of hyperpolarized gas and proton MRI. NMR Biomed. 27, 1461–1467. https://doi.org/10.1002/nbm.3187 (2014).
Stewart, N. J., Norquay, G., Griffiths, P. D. & Wild, J. M. Feasibility of human lung ventilation imaging using highly polarized naturally abundant xenon and optimized three-dimensional steady-state free precession. Magn. Reson. Med. 74, 346–352. https://doi.org/10.1002/mrm.25732 (2015).
Tahir, B. A. et al. Spatial comparison of CT-based surrogates of lung ventilation with hyperpolarized helium-3 and Xenon-129 gas MRI in patients undergoing radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 102, 1276–1286. https://doi.org/10.1016/j.ijrobp.2018.04.077 (2018).
Biancardi, A. et al. in Joint Annual Meeting of the International Society for Magnetic Resonance in Medicine 2018.
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M. & Maier-Hein, K. H. in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. (eds Alessandro Crimi et al.) 234–244 (Springer).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In IEEE International Conference on Computer Vision (ICCV 2015) 1502. https://doi.org/10.1109/ICCV.2015.123 (2015).
Kingma, D. & Ba, J. Adam: A method for stochastic optimization. In International Conference on Learning Representations (2014).
Manjon, J. V., Coupe, P., Marti-Bonmati, L., Collins, D. L. & Robles, M. Adaptive non-local means denoising of MR images with spatially varying noise levels. J. Magn. Reson. Imaging 31, 192–203. https://doi.org/10.1002/jmri.22003 (2010).
Gibson, E. et al. NiftyNet: a deep-learning platform for medical imaging. Comput. Methods Programs Biomed. 158, 113–122. https://doi.org/10.1016/j.cmpb.2018.01.025 (2018).
Abadi, M. et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467 (2016).
Kirby, M. et al. Hyperpolarized 3He magnetic resonance functional imaging semiautomated segmentation. Acad. Radiol. 19, 141–152 (2012).
Kanungo, T. et al. An efficient k-means clustering algorithm: Analysis and implementation. IEEE Trans. Pattern Anal. Mach. Intell. 24, 881–892 (2002).
Tomasi, C. & Manduchi, R. in Computer Vision, 1998. Sixth International Conference on. 839–846 (IEEE).
Li, B. N., Chui, C. K., Chang, S. & Ong, S. H. Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Comput. Biol. Med. 41, 1–10 (2011).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302. https://doi.org/10.2307/1932409 (1945).
Beauchemin, M., Thomson, K. P. B. & Edwards, G. On the Hausdorff distance used for the evaluation of segmentation results. Can. J. Remote. Sens. 24, 3–8. https://doi.org/10.1080/07038992.1998.10874685 (1998).
Shapiro, M. D. & Blaschko, M. B. On hausdorff distance measures. Computer Vision Laboratory University of Massachusetts Amherst, MA 1003 (2004).
Biancardi, A. M. & Wild, J. M. in Medical Image Understanding and Analysis. (eds María Valdés Hernández & Víctor González-Castro) 804–814 (Springer, 2017).
Stewart, N. J. et al. Comparison of (3) He and (129) Xe MRI for evaluation of lung microstructure and ventilation at 1.5 T. J. Magn. Reson. Imaging 48, 632–642. https://doi.org/10.1002/jmri.25992 (2018).
Kirby, M. et al. Hyperpolarized 3He and 129Xe MR imaging in healthy volunteers and patients with chronic obstructive pulmonary disease. Radiology 265, 600–610. https://doi.org/10.1148/radiol.12120485 (2012).
Astley, J. R., Wild, J. M. & Tahir, B. A. Deep learning in structural and functional lung image analysis. Br. J. Radiol.. https://doi.org/10.1259/bjr.20201107 (2020).
Mukesh, M. et al. Interobserver variation in clinical target volume and organs at risk segmentation in post-parotidectomy radiotherapy: Can segmentation protocols help?. Br. J. Radiol. 85, e530-536. https://doi.org/10.1259/bjr/66693547 (2012).
Balachandar, N., Chang, K., Kalpathy-Cramer, J. & Rubin, D. L. Accounting for data variability in multi-institutional distributed deep learning for medical imaging. J. Am. Med. Inform. Assoc. 27, 700–708. https://doi.org/10.1093/jamia/ocaa017 (2020).
Acknowledgements
This work was supported by Yorkshire Cancer Research (S406BT), National Institute of Health Research (NIHR-RP-R3-12-027) and the Medical Research Council (MR/M008894/1). The authors also acknowledge access to high-performance computing equipment available via an Engineering and Physical Sciences Research Council Equipment Grant (EP/P020275/1) and support provided for this equipment grant via Research Software Engineering Sheffield.
Author information
Authors and Affiliations
Contributions
J.R.A., J.M.W. and B.A.T. made substantial contributions to the conceptualization of the work. A.M.B., P.J.C.H., H.M. L.J.S., G.J.C., J.A.E., N.D.W., M.Q.H., J.M.W. and B.A.T. were involved with patient recruitment, image acquisition and/or analysis. J.R.A. performed the deep learning experiments, interpreted data, and conducted statistical analyses. J.R.A. drafted the manuscript. B.A.T. substantively revised the manuscript. All authors reviewed and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Astley, J.R., Biancardi, A.M., Hughes, P.J.C. et al. Large-scale investigation of deep learning approaches for ventilated lung segmentation using multi-nuclear hyperpolarized gas MRI. Sci Rep 12, 10566 (2022). https://doi.org/10.1038/s41598-022-14672-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-14672-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.