Abstract
Based on phylogenetic analyses, strain M2a isolated from honey, an unexpected source of acinetobacters, was classified as Acinetobacter lwoffii. The genome of this strain is strikingly crowded with mobile genetic elements. It harbours more than 250 IS elements of 15 IS-families, several unit and compound transposons and 15 different plasmids. These IS elements, including 30 newly identified ones, could be classified into at least 53 IS species. Regarding the plasmids, 13 of the 15 belong to the Rep-3 superfamily and only one plasmid, belonging to the “Low-GC” family, possesses a seemingly complete conjugative system. The other plasmids, with one exception, have a mobilization region of common pattern, consisting of the divergent mobA/mobL-family and mobS-, mobC- or traD-like genes separated by an oriT-like sequence. Although two plasmids of M2a are almost identical to those of A. lwoffi strains isolated from gold mine or Pleistocene sediments, most of them have no close relatives. The presence of numerous plasmid-borne and chromosomal metal resistance determinants suggests that M2a previously has also evolved in a metal-polluted environment. The numerous, possibly transferable, plasmids and the outstanding number of transposable elements may reflect the high potential of M2a for rapid evolution.
Similar content being viewed by others
Introduction
Acinetobacter genus belong to the γ-Proteobacteria, Pseudomonadales order and Moraxellaceae family and includes aerobic, Gram-negative, catalase-positive and oxidase-negative coccobacilli1,2. The genus has undergone drastic changes before the proposal of Baumann et al.3 was accepted by the subcommittee on Moraxella and allied bacteria4. This proposal limited the genus to oxidase-negative strains, and currently includes ca. 60 validly named species (LPSN, http://www.bacterio.net/-allnamesac.html)5. The taxonomy of the genus relies on classical microbial6 and diverse biochemical and molecular methods. Over the past decades a variety of methods have been used to classify and identify Acinetobacter species, such as DNA-DNA hybridization, AFLP analysis7, amplified ribosomal DNA restriction analysis8,9, sequence analysis of 16S rDNA and several housekeeping genes (gyrB, rpoB, cpn60, fusA, gltA, pyrG, recA, rplB)10, MALDI-TOF mass spectrometry and evaluation of genomic data2. Descriptions of novel species are generally accompanied by a comprehensive set of physiological and nutritional tests, originally developed by11 and later extended by Nemec et al., e.g.12,13,14.
Acinetobacter includes species of different life-styles, from free-living saprophytes to human- and animal-pathogens2,15. Acinetobacter species occur in diverse natural and artificial environments such as forest and agricultural soils, animal and human skin and gut, fresh- and seawater, or even sewage and activated sludge1. Some of them are able to tolerate extreme conditions, for instance low temperature, hydrocarbon-contaminated sites and high osmotic conditions. Despite their high prevalence in most environments, the distribution and ecological roles of Acinetobacter species, apart from pathogenic and nosocomial species with clinical importance, are poorly explored16. The most studied Acinetobacter species is the human pathogen A. baumannii17,18, which have attracted exceptional attention because of its pathogenicity and multiresistance19. Less information is available on non-baumannii acinetobacters15, living in a wide range of environments including habitats contaminated with heavy metals20 or oil21,22, cold habitats23,24 or high osmotic environments, such as saltern ponds25 or floral nectar26. To our knowledge, there are no reports of Acinetobacter spp. isolated from honey27, however there are reports on Acinetobacter spp. in the honey bee gut28. Acinetobacter was suggested as a model organism in the environmental microbiology and pathogenesis2,29,30 due to its ecological and clinical importance, the utilization of various kinds of carbon sources and sufficient growth on simple media and the environmental characteristics, which substantially differ from those of the most common enteric model organism, E. coli. Several Acinetobacter spp. have been regarded as potentially important microorganisms in both environmental and biotechnological applications like bioremediation of soil and water, or production of “bioproducts” (polysaccharides, polyesters, enzymes)31.
Mobile genetic elements (MGEs) often harbour various kinds of genes endowing their hosts with resistance to antibiotics and heavy metals, or beneficial traits like virulence or metabolism of unusual substrates32,33,34. Well-known vehicles of such genes are plasmids often capable to transfer horizontally, even between unrelated bacterial species by conjugation or natural transformation. Plasmids therefore play an important role in the evolution and adaptation of bacteria. Even though many resistance genes are not located on the resistance islands but are scattered over the genomes, the genomic resistance islands and plasmids are key players in the emergence of antibiotic resistant Acinetobacter strains, which represent a significant health threat e.g. the nosocomial pathogen A. baumannii19,35,36,37. More frequent occurrence of plasmids was observed in 75 clinical Acinetobacter isolates classified into four non-baumannii species and three different multiresistance patterns. Plasmid DNA fingerprinting showed that >84% of these strains contained up to 15 plasmids38. In a comparative study, A. lwoffi isolates were found to carry more plasmids (≤20) than A. anitratus (≤8)39. Resistance genes are often associated with transposons and integrons19,35,40,41,42,43, in addition non-integron cassette streptomycin/spectinomycin resistance gene aadA27 was also observed in plasmids identified in ancient (isolated from permafrost) and recent environmental Acinetobacter isolates44.
In this study we describe an Acinetobacter lwoffii strain named M2a that was isolated from a Transylvanian honey sample and proved to carry an outstanding number of MGEs. The aim of this project was to investigate the bacterial community of honey and the intestinal tract of honeybees derived from a nearly natural rural meadow, and to isolate Lactobacilli or other species that might have probiotic traits. The different stains obtained were classified based on phylogenetic analysis of their 16S rRNA gene, and their plasmid content was also examined. Among these isolates, M2a appeared to be interesting as Acinetobacter spp. are rarely isolated from sugar-rich environments and the preliminary examination suggested that the strain carry numerous, possibly undescribed, plasmids.
Results and Discussion
Isolation and characterization of Acinetobacter strain M2a
Strain M2a was isolated from a honey sample derived from a nearly natural meadow in Transylvania together with many other yet uncharacterized Acinetobacter, Lactobacillus, Lysinibacter, Saccharibacter45 and Sphingobacterium46 strains that could grow under conditions favourable mostly for lactobacilli. Thus, M2a was isolated from an MRS + CaCO3 agar plate incubated under CO2-enriched condition at 35 °C for 48 h. It proved to be catalase-positive whilst negative in methyl red, Voges-Proskauer, indole, citrate utilization, urease and oxidase tests. When grown on TSI agar, the strain proved to be a glucose, lactose and sucrose non-fermenter, and did not produce detectable amount of H2S or other gases. Optimal growth occurred at 30 °C on LB agar, while slower growth was observed at 37 °C, and no growth occurred at 44 °C.
BLAST search using the PCR-amplified 16S rDNA as a query sequence suggested that M2a can be classified as an Acinetobacter sp.. The preliminary analysis of its plasmid content indicated that the strain carries multiple small and medium size (<20 kb) plasmids (Suppl. Fig. S1). Due to the fact that Acinetobacter strains have rarely been isolated from sugar-rich environments like honey28 and to the diverse plasmid content found in M2a, we decided to investigate its genome by WGS.
The whole genome sequence
3.6 million Illumina MiSeq reads representing ca. 76× coverage of the whole genome were de novo assembled using A5-miseq and the resulting 277 scaffolds (average length: 13053 bp, median: 5605 bp) were annotated by the RAST server. 3637 annotated genes, 153 tRNAs and 36 rRNAs were identified in the scaffolds, whose total length was 3 615 619 bp with 40.44% GC-content as reported for acinetobacters2. BLAST searches in the GenBank database with the 277 scaffold sequences indicated that of 168 contained chromosomal sequences, 57 showed at least partial homology to known plasmids, while the remaining 52 scaffolds represented different IS element sequences (Suppl. data 1).
Phylogenetic analysis
Phylogenetic relationship of M2a was determined by analyzing the 16S rRNA gene and housekeeping genes rpoB, gyrB and recA (Suppl. data 2). The phylogenetic tree based on the 16S rDNA sequences of M2a and representative members of the family Moraxellaceae (Suppl. Table S1a) indicated that our isolate belongs to the Acinetobacter genus (Fig. 1a). For more exact classification, WGSs of 44 Acinetobacter species with validated names (Suppl. Table S1b) were downloaded from public databases and phylogenetic trees were generated for their 16S rDNA (Fig. 1b), rpoB, gyrB and recA genes (Suppl. Fig. S2). A. lwoffii was found to be the closest relative of our isolate and this was also supported by analyses of further housekeeping genes, such as dnaJ, groEL and gyrA (data not shown). A recent study revised the taxonomy of strains formerly classified as A. lwoffii, genospecies GS8 and GS9, and created a new species, A. pseudolwoffii for group GS814. To decide which species M2a belongs to, an rpoB-based analysis was carried out for 13A. lwoffii and 13A. pseudolwoffii strains as described previously by14. The rpoB tree confirmed that M2a is a strain of A. lwoffii (Suppl. Fig. S3).
Phylogenetic relationship of strain M2a. (a) Phylogenetic tree for the 16S rDNA sequences of representative species from genera of Moraxellaceae including strain M2a. (b) Phylogenetic tree for the 16S rDNA sequences of 44 species of the genus Acinetobacter including strain M2a. The 16S rDNA sequence of Moraxella lacunata was included as outgroup. Trees were generated using the neighbour-joining method. Bar, 0.01 changes per nucleotide position.
Antibiotic and metal resistance determinants
Antibiotic susceptibility tests indicated that M2a had low level ampicillin resistance (<50 μg/ml Ap), but was sensitive for antibiotics such as ciprofloxacin, chloramphenicol, erythromycin, florphenicol, gentamycin, kanamycin, nalidixic acid, neomycin, rifampicin, spectinomycin, streptomycin, tetracycline and zeocin in the concentrations used in standard microbial methods (Table 1). Although the MIC value of M2a was determined as 100 μg/ml for ampicillin (Ap), the 100× dilution of the tested overnight culture (containing ca. 1.25 × 104 cells) gave only a few colonies on LB agar + 100 μg/ml Ap. Additionally, the CFU was 2 orders of magnitude lower in the presence of only 50 μg/ml Ap than in absence of Ap (1.0 × 106 vs. 2.5 × 108/ml, respectively), indicating that less than 0.5% of M2a cells could form colonies on LB agar + 50 μg/ml Ap. A similar gradual increase of CFU was observed with decreasing concentrations of ciprofloxacin and rifampicin.
After the susceptibility test, the WGS of M2a was searched for potential resistance genes using public databases CARD, MEGARes and the ARG-ANNOT server. 7 unspecific AR loci and a gene for carbapenem-hydrolyzing class D β-lactamase OXA-283 (OXA-134 family) were identified in the chromosome (Table 2). BLAST searches revealed that bla OXA-283 of M2a is identical to bla OXA of Acinetobacter sp. CIP A162 (NG_049581) and also occurs in many A. lwoffii strains. It does not carry the 9-bp deletion characteristic for several strains of A. lwoffii/genomic species 947 and shows 93–99% similarity to its homologs in A. lwoffii. Thus, the presence of the chromosomal OXA-134 gene further supports the results of the phylogenetic analyses, which advocated that M2a should be designated as A. lwoffii. The best match was found with A. lwoffii strain ZS207, where the orthologous gene carries only 3 SNPs (causing no amino acid changes) and the 400 bp upstream regions are also identical. Despite the apparently intact coding sequence and upstream region (probably containing the promoter), M2a proved to be sensitive to >50 μg/ml ampicillin. Disc tests were performed with new generation carbapenems cefotaxime and ceftazidime, against which the OXA-134 family β-lactamases are efficient enzymes48. The inhibitory zone diameter was >26 mm for each drug indicating that M2a is sensitive to these antibiotics, similarly to other A. lwoffii strains carrying chromosomal OXA-134 genes48.
The M2a genome proved to also carry many metal resistance determinants (Table 2), thus the resistance of M2a for several heavy metal ions was assayed. MIC values were similar to those of A. lwoffii strains isolated from Kolyma Lowland permafrost, while the Hg-resistance proved to be outstanding (>0.075 mM, Table 1). This broad range of metal resistance raises the possibility that M2a might have evolved in a metal-polluted environment.
Mobile genetic elements in M2a
Screening for integrons
Resistance determinants are often associated with mobile genetic elements (MGE) such as plasmids, transposons and integrons34. Thus the WGS of M2a was thoroughly screened for such elements. Fourteen integrase-like genes encoding putative site-specific recombinases were found in the annotated scaffolds (Suppl. data 3), but all appeared to be related to phage integrases and none to integron integrases. BLAST searches for sequences of primer pairs designed to detect genes for IntI classes 1 to 349 and class 1 integron cassettes50 were also negative, indicating that M2a genome does not carry integrons.
Characterization of transposable elements
In contrast to integrons, a large number of IS elements were found in the M2a genome by BLAST searches against the IS Finder database and additional, yet unidentified elements were discovered during the thorough analysis of scaffold termini. 46 of the 277 scaffolds represented full length or partial ISs without flanking sequences, and 200 of the remaining 231 scaffolds ended in IS sequences at either (38 scaffolds, 16.5%) or both termini (162 scaffolds, 70.1%). Due to the fragmentation of the WGS, the number of IS elements could not be determined by simple counting (see Suppl. Methods). According to our estimation, 256 ISs, including 201 full length and 55 incomplete copies, occur in the chromosome and plasmids of M2a. No IS elements were identified with 100% identity to ISs available in IS Finder database. The divergence from the closest relatives in the database ranged from 99.8% DNA similarity to the level of homology undetectable by MegaBLAST comparison. In the latter cases, the new elements were classified according to the protein sequence of their putative transposase. The ca. 256 elements represent more than 55 IS species from 15 different families (Suppl. Table S2a). The exact number of IS species was hard to determine as in M2a we found many sequence variants of closely related elements that could be classified as different species or iso-elements (iso-ISs) depending on their sequence divergence. Unfortunately, there is no widely accepted cut-off value of nucleotide sequence identity for separation of IS species. IS Finder attributes new names to ISs when the similarity for protein and DNA sequences is <98% or <95%, respectively (P. Siguier, pers. comm.), and this practice was adopted in this work. Finally, 22 newly identified full length ISs have been named and submitted to IS Finder and many further IS sequences, mostly incomplete or frameshift-mutant elements, were identified as new ISs without designation (highlighted by red in Suppl. Table S2a).
IS families and transposons in M2a
Several IS families are represented by a single IS copy, such as families IS21, IS256, IS481 and IS1595, or by only few copies, like families IS6 and ISNCY. In contrast, IS30 and IS701 families are also represented by one IS species (isoISAba125 and isoISAba11, respectively), but with more than 10 copies. The other seven families include at least three different IS species and often more than 30 copies.
The 35 IS1-family elements could be classified into three IS species: isoISPa14 and other two types that are only 71% similar to each other and highly divergent from ISPa14. Thus, the latter two were assigned as new IS1-family elements, ISAlw2 and ISAlw3.
In M2a ca. 10 IS3-family ISs with more than 40 copies were found. All four subfamilies were represented and six new IS species could be identified. The most abundant ISs belonged to isoISAba14 and the isoISAba18/19/29/34 complex. These ISs are present in several different variants in the M2a genome and show 3–12% divergence from their closest known relatives. Out of the four fully assembled IS51-subfamily copies, the partial left and right end sequences found at scaffold termini could not obviously be paired due to different levels of their homology to the prototype ISs. Thus, the number of different IS species could not be exactly defined in the isoISAba18/19/29/34 group (Suppl. Table S2a).
IS4 is the next dominant family represented by 42 copies of seven IS species. 21 copies are iso-elements of ISAba1, ISAba33 and ISAbe18 and are almost identical to their prototype. The other copies belong to four completely new IS species (ISAlw7-ISAlw10). ISAlw8 and ISAlw9 copies form two slightly divergent sub-types, while ISAlw10 copies are uniform.
The ISs with the most copies (55) in M2a genome belong to the IS5 family, which is represented by all three subfamilies and 12 IS species. Besides the most abundant element, isoISAba31, four new members of the IS427 subfamily were discovered. ISAlw11 shows marginal homology to ISAbe13 and could only be classified according to its transposase protein sequence, as well as the two different ISPssp5-related elements, ISAlw12 and ISAlw13. Four different ISs represent the IS903 subfamily, three of them are closely related to IS17, ISAha2 and ISAba12, respectively, while a truncated element appeared to be a new IS related to ISAba40. Three further elements were classified into the ISL2 subfamily. In addition to the slightly divergent copies of isoISAba27, two new IS species, ISAlw14 and ISAlw15, distantly related to ISCaa2 and ISCaa3, respectively, were identified (Suppl. Table S2a).
M2a carries 12 IS66-family IS copies. Except isoISAba17, isoISAba25 and the newly identified ISAlw16 and ISAba49-related elements, the other copies could not confidently be classified, as their left and right parts show different levels of homology to ISAba25, ISAba46 and ISAba49. These elements differ not only from the prototype ISs but also from each other, and they represent at least five variants.
The next family, IS630, is represented in M2a by three species. Besides isoISAba44, two new elements were identified. ISAlw17 is a distant relative of ISAba44, while ISAlw18 is distantly related to ISMae24. Two types of ISAlw18 occur in the chromosome: the right inverted repeat (IRR) of one copy differs at 3 positions from that of the other four identical copies. Interestingly, the single iso-element with ‘divergent’ IRR is more prevalent in other Acinetobacter strains, e.g. A. lwoffii ZS207 and A. wuhouensis WCHAW010062 (GenBank CP033133.1), harbouring three and 28 identical copies, respectively. Moreover, slightly different ISAlw18 copies with the same ‘divergent’ IRR are also present in several A. lwoffii plasmids (pALWEK1.1, pALVED3.6).
Finally, 16 IS982-family elements were also found: IsoISAba9 and isoISAba825 copies are almost identical to the prototype elements, while isoISAcsp2 copies show larger divergence (87–96% similarity). Furthermore, two new family members, ISAlw19 and ISAlw20 were discovered. Based on their transposase protein sequence, their closest relative is ISNeu1, although their DNA sequences are very different.
In addition to the IS elements, three different transposons were identified in M2a. Besides the incomplete TnAs2-related Tn3 family transposon, carrying the mercuric resistance (mer) module (Table 2), a Tn7-related element was found in the chromosomal scaffold sc_30. Although the termini of this element could not be exactly determined, the presence of a complete gene set (tnsABCDE) characteristic for Tn7 transposons and the occurrence of close (90–95% similar) relatives of this Tn in several Acinetobacter strains (A. schindleri SGAir0122, A. johnsonii IC001) suggest that it is an intact transposon. In addition, a compound transposon, named as Tn6682, consisting of two directly oriented identical isoISAba14 copies bracketing an alkyl sulfatase and a tetR-family regulator gene was also identified. The same 5.5 kb transposon (with 100% identity) occurs in A. ursingii M3 (AP018824.1), indicating a recent interspecific lateral transfer event. Further compound transposon-like structure was found in sc_211, where two inversely oriented isoIS125 copies surround an ORF encoding a protein of unknown function. Since a similar transposon-like unit does not occur in GenBank entries, there is no indication of its transposition.
For comparison, similar analysis was carried out for A. lwoffii ZS207, which appeared to be the closest sequenced relative of M2a. Strain ZS207 proved to carry a similar set of ISs, but the copy number was roughly half of what we found in M2a (Suppl. Table S2b). There are 14 common IS families of the two strains, although M2a contains a new IS1595-family element (ISAlw21) that is missing from ZS207, but lacks IS200- and ISL3-family elements. Altogether, 123 copies of 51 IS species were found in the chromosome and plasmids of ZS207. 12 ISs newly identified in M2a and an incomplete copy of the new Tn7-like transposon are also present in ZS207. 9 further new ISs were identified, some of them appeared to be distant relatives of several new elements found in M2a. In general, the copy number of ISs is lower in ZS207 than in M2a. While the maximum copy number in ZS207 is 8 and most elements occur in less than five copies, M2a has seven ISs with 10–22 copies and 25 ISs occur in at least five copies.
A similarly high number of IS elements has been reported in the A. baumannii strain SDF isolated from body lice, but in contrast to the remarkable diversity of ISs of M2a, its IS population exclusively contains hundreds of ISAba6 and ISAba7 copies. This might have important role in genome reduction of their host by recombination and gene disruptions51.
Plasmids of strain M2a
Identification of the different replicons
Like some other A. lwoffii strains isolated from permafrost or arsenic-polluted environments, such as strains ED9-5a, ED23-35, ED45-23, EK30A52 or ZS207, M2a also proved to carry multiple plasmids (Suppl. Fig. S1). Many different plasmid sequences identified mostly in Acinetobacter strains were retrieved from GenBank by BLASTn search with the 57 plasmid-related scaffolds found in the WGS (Supplementary data 1). By screening the WGS for plasmid-related genes, such as genes for replication initiation (rep) and conjugal transfer (tra, mob), 15 plasmids could be identified (Table 3, Fig. 2), which appears to be exceptional compared to the mentioned strains that have 8–12 plasmids52, (ZS207: CP019144 to CP019152).
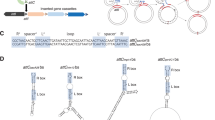
Schematic maps of plasmids identified in M2a. The colour code is: red, rep region (oriV, repB, DBP gene); green, mobilization; blue, TA systems; yellow, IS elements; purple, cargo genes. The related segments of pAVAci130 and pAVAci 144 are shown by grey. The regions of pAVAci84 homologous to pZS-13 and pZS-6 are indicated above the plasmid map by brown and light green bars, respectively.
Eight plasmid sequences (pAVAci14, pAVAci84, pAVAci117, pAVAci119, pAVAci130, pAVAci144, pAVAci145 and pAVAci147) were assembled into single scaffolds by the A5-assembly. Their sequence could be circularized based on their overlapping end sequences and sealing PCRs carried out with appropriately designed primers facing outward of the ends of the respective scaffold. Further three plasmid sequences (pAVAci94, pAVAci116 and pAVAci176) could be completed by manual assembly of scaffolds based on their overlapping end sequences. In these cases PCR verification of the assembly and, if required, sequencing of the sealing PCR fragment were also accomplished. To confirm that these sequences are circular extrachromosomal elements, they were cloned (except the 46 kb pAVAci14) in an R6K-based E. coli plasmid using unique restriction sites found in the plasmid sequences (see Methods). These clones, maintained in TG2 λpir strain, were also used to test whether the plasmids are able to replicate in E. coli. The cloned pAVAci plasmids were introduced into TG1 cells, which do not support the maintenance of the R6K-based replicon of the cloning vector. None of them resulted in colonies indicating that these plasmids, similar to most Acinetobacter plasmids, are unable to replicate in E. coli.
Four additional plasmids (pAVAci98, pAVAci115, pAVAci127 and pAVAci167) were identified based on their rep and other genes characteristic for plasmids (par, mob, toxin-antitoxin (TA) module)53. These plasmid sequences could not be completed even by manual assembly due to the high number of scaffolds ended with similar IS elements. The replicon regions of pAVAci98 and pAVAci127 are similar to large plasmids like pALWEK1.1, pmZS and pALWED2.1, which all carry numerous IS elements as well. However, many scaffolds represent as yet unknown sequences, which prevented the full assembly of the sequences using the published relatives as reference sequences.
General features of plasmids in M2a
Despite the large diversity of plasmids in M2a, some common features could be seen in their replication and mobilization regions (Fig. 2). All but two plasmids have repB gene coding for a Rep-3 superfamily replication initiation protein. In most cases, repB is followed by a putative DNA-binding protein (DBP) gene as was found in many other Rep-3 superfamily replicons37. The noncoding upstream region of repB always contains four to nine directly oriented imperfect or perfect repeats of about 20-bp motif. This part of the plasmids possibly functions as the iteron region of the replication origin (oriV). The iterons are often accompanied by shorter direct or inverted repeats (DR or IR, respectively) probably belonging to the entire functional oriV. The two exceptions are pAVAci130 and pAVAci144 that have no repB gene and where an iteron region could not be found.
The other generally occurring plasmid-related function was the mobilization genes (mob). Although only pAVAci14 appears to have a complete gene set for conjugal transfer, all but one other of the plasmids carry a mob region containing plasmid mobilization genes like mobA/mobL and mobC, mobS or traD. The common pattern of mob regions are the divergently oriented mobA/mobL-family nickase/relaxase gene and a short mobS-, mobC- or traD-like gene, which are separated by about 200–300 bp non-coding sequence. The localization between divergent mob genes and the presence of an array of IR motifs suggest that these non-coding regions contain the origin of transfer (oriT)54,55. Since these plasmids have no other transfer related genes such as genes for Type IV pilus components and assembly factors or coupling protein, it is more possible that they can be trans-mobilized by other conjugative plasmids rather then they are self-transmissible. Although, the occurrence of similar mob regions in Rep-3 superfamily plasmids is not extraordinary, their horizontal transfer has yet to be demonstrated37. The phylogenetic trees generated for the RepB and the MobA protein sequences (Fig. 3) differ significantly, which indicates that the replicons and the mobilization regions evolved mostly independently of each other and intensive reshuffling might occur between the different plasmids by recombination and transposition.
Characterization of the M2a-derived plasmids
The only exception to the pattern described above for mob regions can be seen in pAVAci14, where the putative oriT-like region was found between the directly oriented parA and mobC genes (Fig. 2). The closest relative of the 45.7 kb pAVAci14 is pHHV35 (Table 3) belonging to the “Low-GC” group of plasmids56, where oriT have been localized at the same position as predicted in pAVAci14. Besides the replication (repB, oriV), maintenance (parAB,) and transfer functions, pAVAci14 contains relatively few accessory genes (Type III restriction/modification system, umuCD-like repair genes, dnaJ, a Ser-recombinase family resolvase, an MFS-1 family membrane protein, a TPR-repeat-containing protein, several hypothetical genes and an ISAcsp5 copy). Interesting dissimilarity to pHHV35 backbone is that pAVAci14 contains additional mobC and mobA/mobL mobilization genes encoded by partially overlapping ORFs in a single operon. These genes along with seven other ORFs are inserted amongst traB and oriV interrupting the conserved backbone (Fig. 2). Although pAVAci14 appears to have no resistance determinants, the group of conjugative Low GC plasmids have been suggested to be important factors in environmental spread and interspecies transfer of antibiotic resistance between bacterial communities of manure and soil56,57,58.
Plasmids pAVAci130 and pAVAci144 devoid of rep genes are highly homologous to plasmids pALWEK1.6/pZS-7 and pZS-8 (Table 3). pAVAci130 contains only one or two base substitutions compared to the published pALWEK1.6 and pZS-7 sequences, respectively, while pAVAci144 differs from pZS-8 at 134 positions (2.8%) and lacks the IS5 family element inserted in pZS-8 near mobA/mobL. The two plasmids are related to each other as their mobilization regions and a 1.6 kb other segments with a hypothetical gene and a 0.9 kb non-coding sequence, which possibly includes the replication origin, show 88% and 84% similarity, respectively (Fig. 2). A ca. 670 bp tract of the non-coding regions shows 77% homology to the upstream region of orf2 of pRAY, a representative of plasmids also lacking rep gene and widely distributed in Acinetobacter59. The almost identical sequence of the three plasmids, pAVAci130, pZS-7 and pALWEK1.6, which were found in different strains of A. lwoffii isolated at different time and locations, suggests recent horizontal transmission of this plasmid, which indirectly supports the assumption of its ability for trans-mobilization. Interestingly, pALWEK1.6 derives from A. lwoffii strain EK30A that was isolated from permafrost of 1.6–1.8 million-year-old Pleistocene sediment52, which might suggest that the evolution of this plasmid is exceptionally slow (1 base substitution/1.6 M year) or the strain EK30A was a recent environmental contamination in the permafrost sample.
Plamsid pAVAci84 appears to be the fusion product of two plasmids as it carries two complete rep and mob regions and two TA systems. The RepB proteins are closely related (Fig. 3a) and the putative oriVs are also similar. Both carry an array of a 9-bp AT-rich direct repeats, however, the following array of four 22-bp repeats (the iterons) are different. Both mob regions consist of divergent mobC and mobA genes and an oriT-like sequence between them. MobA proteins are also closely related, however, similarly to the RepB-s, they are not the closest relatives in M2a (Fig. 3b). The presumed fusion of the two plasmids resulting in pAVAci84 seemingly occurred between the putative oriT regions of the parental plasmids. One component of pAVAci84 (1–2409 and 9769–14155 bp) is almost identical to pZS-6. The only differences are 3 SNPs and the duplication of a 206-bp tract downstream of the Fe-dioxigenase gene. The other component, however, is more closely related to pZS-13, from which it differs only in 6 SNPs and in the presence of an AAA-family ATPase gene inserted between the brnT-family toxin gene and the rep region (Fig. 2). The sequence of pZS-6 and pZS-13 contains several regions showing 76–82% similarity, but one of the longest identical sequences occurs in their putative oriTs, which might explain that the fusion took place at this region. Although the formation of pAVAci84 can be explained by homology-dependent recombination, the involvement of the putative relaxases in this process cannot be excluded.
The 17-kb pAVAci94 appears to be a new plasmid species. Its rep region, consisting of oriV, repB and a putative DBP gene, and the brnA-brnT TA system show 92% and 88% homology to the respective regions of the A. baumannii plasmid pO237-4. In contrast, the mob region is 88% similar to that of pALWED3.2 (Table 3). Additional parts of pAVAci94 (from yfgC to a group I intron nuclease-domain-containing protein gene) has no homologs among DNA entries of GenBank.
The 13-kb pAVAci116 also seems to be a new plasmid species with mosaic structure. Its rep region (oriV, repB and a DBP gene ylxM) is similar to that of pMS32-3, however, the mob region (mobA-oriT-traD), the TA system and a sulphate permease gene are related to those of pOXA58-AP_882 (Table 3).
The ca. 9-kb plasmids pAVAci117 and pAVAci119 have closely related mob regions, while their rep regions are more dissimilar (Fig. 3). The rep region of pAVAci117 is only partially homologous to several A. baumannnii plasmids (e.g. pAb825_36 or p597A-14.8), while that of pAVAci119 is 82–85% homologous to other A. baumannnii plasmids such as p2ABSDF or pEH_gr3. Almost half of pAVAci117, coding for three cargo genes, has no homologs in the database.
The two smallest plasmids, pAVAci145 and pAVAci147 do not carry accessory modules besides the basic replicon and the mob region (Fig. 2). The putative DBP gene generally located downstream of repB is missing from both plasmids and their RepBs, and the mob regions are quite different (Fig. 3). The mob region of pAVAci145 consists of mobC-oriT-mobA, while the other plasmid has a traD-like gene instead of mobC and their MobA proteins are also located on different branches of the phylogenetic tree (Fig. 3b).
The last small plasmid, pAVAci176 also lacks a DBP gene, however the ORF downstream of repB might have similar function. The mob region corresponds with the mobC-oriT-mobA pattern and the cargo module, except the ORF-3-like protein gene, is related to that of pALWEK1.4.
Among the remaining plasmid-related scaffolds, representing 276.8 kb sequence, four were found that carried similar rep and mob regions identified in the fully assembled plasmids. The scaffold sc_98 contained a complete Rep-3-type replicon (oriV-repB-DBP gene) and a mob region (mobS-oriT-mobA). Two large scaffolds could unambiguously be joined to sc_98 and the resulting sequence carried three TA-systems and a gene set for resistance to heavy metals (Fig. 2). A BLAST search with this segment as query indicated that this plasmid, designated as pAVAci98, is most closely related to the large plasmid pALWEK1.1 which also carries metal resistance modules.
The next plasmid-derived scaffold was sc_115, which carried two different replicons and a mobS-oriT-mobA-type mob region. The two replicons are not closely related as one of the RepB proteins is closer to RepB of pAVAci145, while the other forms a distant branch with those of pAVAci84, pAVAci94, pAVAci98 and pAVAci176. The rep_1 region consists of repB and oriV assembled from five 11-bp repeats followed by nine 23-bp iteron repeats, while rep_2 includes an additional HTH-17 type DBP gene and the oriV contains four 22-bp iteron repeat preceded by a complex array of IRs. Accordingly, the closest relatives of rep_1 + mob and rep_2 regions were found in different plasmids, i.e. pRGFK1137 and pIC001C, respectively (Table 3). The backbone of this plasmid, named as pAVAci115, also carries a TA-system and a kfrA-like gene, which may participate in plasmid maintenance. Sc_115 could be joined to several short scaffolds, which added some cargo genes and a complex IS-in-IS segment to the backbone (Fig. 2), however it could not be completed.
pAVAci127 was the only plasmid that lacks a mob region. Its basic replicon (repB and oriV, no DBP) and the plasmid partitioning genes parAB are bracketed by tracts of IS elements (Fig. 2). The rep-par segment is almost identical to the homologous parts of the 190–200 kb plasmids pALWED2.1 and pmZS. These plasmids contain many metal-resistance determinants and, similarly to pAVAci127, seem to be not mobilizable.
The last replicon identified in M2a was pAVAci167. Its rep region found in sc_167 contains a DBP gene, repB and the oriV. The mob region shows the traD-oriT-mobA pattern. The closest relatives of pAVAci167 (based on the rep and mob regions) are 13–25 kb plasmids identified in A. baumannii and A. pittii strains (i.e. pB8300), however pAVAci176 carries a ca. 5 kb region containing a TA-system, a resolvase and a methyltransferase gene, which is 95% similar to a segment of pZS-20 (Table 3).
Since pAVAci115 and pAVAci167 are related to smaller (7–30 kb) plasmids, while pAVAci98 and pAVAci127 show similarity to large (~200 kb) plasmids like pALWEK1.1, pmZS or pALWED2.1, it is presumable that most of our unassembled plasmid scaffolds (Suppl. data 1) belong to pAVAci98 and pAVAci127. These parts of plasmids carry lots of IS elements, code for metal resistance efflux systems and many metabolic (oxidases, reductases, membrane transporters) and unknown functions, which may contribute to the adaptability of the host organism.
Conclusions
A. lwoffii strain M2a that was isolated from a Transylvanian honey sample derived from a nearly natural environment proved to contain 15 different plasmids, more than 250 IS elements of 15 IS-families and some unit and compound transposons. Besides several antibiotic efflux systems and an OXA-134 family β-lactamase gene, the strain carries numerous chromosomal and plasmid-borne heavy-metal resistance determinants similarly to A. lwoffi strains isolated from metal-polluted environments or permafrost52. One out of its 15 plasmids, the “Low GC” family plasmid pAVAci14, has an apparently complete conjugative system, while the others, except pAVAci127, have a mob-region showing common pattern. All mob regions consist of a divergently oriented mobA/mobL-family relaxase gene and a mobS-, mobC- or traD-like gene separated by a putative oriT of about 200–300 bp with several IR motifs. The frequent occurrence of such mob regions suggests that these plasmids are trans mobilizable, perhaps by pAVAci14. Plasmid pAVAci130 of M2a are almost identical to plasmids of A. lwoffi strains isolated recently from metal-polluted environments and from 1.6 million-year-old permafrost sediment52. The first case may indicate a recent lateral transfer event, however the second is hard to explain without supposing that pAVAci130/pALWEK1.6 evolved extremely slowly (1 base substitution per 1.6 M years) or that the source strain EK30A was a recent contaminant in the permafrost sample. Regarding A. lwoffii M2a, it was probably a bee-delivered contamination27 in the honey sample it was isolated from. Although M2a does not show extensive antibiotic resistance, its several plasmids are related to factors of environmental spread of AR between bacterial communities56,57,58. The high number of its presumably transferable plasmids and the outstanding number and diversity of IS elements that may be involved in reshuffling the chromosomal and plasmid-borne gene content, may indicate the potential of such strains to rapidly become a multiresistant pathogen60,61,62, which should not be overlooked.
Methods
Isolation of bacteria from honey samples
The project initially aimed to isolate lactobacilli and other bacteria from honey produced in nearly natural environment of Transylvanian meadow near Székelykeresztúr (Cristuru Secuiesc, Romania). The honey samples were collected in 2014 and stored at room temperature until the analysis (isolation of bacteria occurred within two months after sample collection). Approximately 2 g of honey samples were suspended in 15 ml peptone water (0.1% w/v, 0.5% NaCl, pH 7.2), then were centrifuged (10 min, 25 °C, 3000 × g) and the pellet was resuspended in 500 µl peptone water. Twenty-five μl suspension was plated on MRS agar63 with or without 0.8% CaCO3 and incubated for 48 h under CO2-enriched condition (in the presence of 5% CO2) at 35 °C. Strain M2a was isolated from MRS agar + CaCO3 plate. The original colony was streaked twice on LB agar plates and grown under aerobic condition, which appeared more convenient to maintain the strain.
Microbial techniques and biochemical tests
M2a and E. coli strains were grown at 30 °C or 37 °C, respectively, in Luria-Bertani (LB) broth or agar plates and were maintained at −70 °C in LB broth containing 30% glycerol. The MICs for antibiotics and heavy metals for M2a (Table 1) were determined by the agar dilution method64 with minor modifications. Bacteria were grown overnight at 30 °C in LB broth and then the culture was serially diluted 10-fold to 107 × with 0.9% NaCl solution. Five μl of the bacterial suspensions (cc to 107 × dilutions of the 2.5 × 108 CFU/ml culture) was dropped onto LB plates containing different concentrations of the examined antibiotics or heavy metal salts. The used concentrations of antibiotics (μg/ml) were as follows: ampicillin: 50, 75, 100, 125, 150, 175, 200; chloramphenicol: 5, 10, 15; ciprofloxacin: 0.25, 0.5, 075, 1.0, 3.0, 6.0; erythromycin: 25, 50, 75, 100, 125, 150; florphenicol: 10, 15, 20, 25; gentamycin: 2, 4; kanamycin: 10, 15, 20; nalidixic acid: 5, 10, 15; neomycin: 10, 20, 30, 40; rifampicin: 25, 50, 75, 100; spectinomycin: 20, 50, 60, 70, 100, 125, 150, 300; streptomycin: 10, 25, 50; tetracycline: 2, 4; zeocin: 25, 50, 75, 100; and of metal ions (μM): HgCl2 (Hg2+): 15, 30, 45, 60, 75; CdCl2 × H2O (Cd2+): 10, 25, 50, 100, 200, 400, 600, 800, 1000; CoCl2 × 6H2O (Co2+): 10, 25, 50, 100, 200, 400, 600, 800, 1000; CuSO4 × 5H2O (Cu2+): 900, 1800, 2700, 3600, 4500; ZnSO4 × 7H2O (Zn2+): 200, 400, 800, 1600, 3200. Concentration of viable cells (CFU/ml) was determined on LB plates without antibiotics or heavy metal salts. The plates were incubated at 30 °C for 24 h and visually evaluated. MIC value was determined on the 100 × dilution drops (ca. 1.25 × 104 cells/drop).
Biochemical tests e.g. methyl red, Voges-Proskauer, indole, citrate utilization, catalase production, urease, and oxidase tests were performed as described previously65. Glucose, lactose and sucrose fermentation, and gas and H2S production was examined on Triple Sugar Iron agar (Biolab Inc., Budapest, Hungary). Growth was tested at 30/37/44 °C for 1 days on Luria–Bertani (LB) and Eosin Methylene Blue agar (Biolab Inc., Budapest, Hungary).
DNA purification and sequencing
Total DNA for WGS was purified from M2a using Qiagen Blood & Cell Culture Kit with Genomic-tip 20/G (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA quality and quantity were tested on Ethidium-bromide-stained agarose gel (1% agarose, 1% TBE buffer) and NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The 600-630-bp fragment library was prepared by UD GenoMED (Debrecen, Hungary) and 2 × 300-bp paired-end genome sequencing was performed by University of Szeged, Department of Biochemistry and Molecular Biology (Szeged, Hungary) as a custom service using Illumina’s MiSeq platform.
For cloning and PCRs plasmid DNA of M2a was extracted from 100 ml culture using the QIAGEN Plasmid Midi Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Cloning procedures and transformation of CaCl2 competent E. coli cells were carried out according to66. M2a-derived plasmids were cloned in the pir-dependent vector pSG76C67 and maintained in E. coli strain TG2 λpir, a derivative of TG266 obtained by lysogenization with λpir phage isolated from S17-1 λpir strain68. Colony PCRs were performed using Dream Taq polymerase (Thermo Fisher Scientific) as described previously69. Oligonucleotide primers used in this work are listed in Suppl. Table S3. Sanger sequencing was carried out on ABI Prism 3100 (Perkin Elmer) by BIOMI Ltd. (Gödöllő, Hungary). The bacterial isolates were first classified by sequencing of their 16S rDNA segment amplified in colony PCR using primers 27for and 1492rev70.Thermal cycling was as follows: initial denaturation at 96 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 90 s and a final extension at 72 °C for 7 min.
Bioinformatics
The Illumina MiSeq reads were de novo assembled into scaffolds using A5-miseq pipeline71. The scaffolds were annotated using the RAST server72. The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VCND00000000. The version described in this paper is version VCND01000000. The scaffolds of WGS available in GenBank was re-annotated by the NCBI’s annotation server. The completed plasmid sequences identified in M2a have also been deposited at GenBank under the accession numbers listed in Table 3.
All homology searches were carried out using BLAST73 in the NCBI database (http://www.ncbi.nlm.nih.gov/). Alignment of M2a scaffolds to the reference A. lwoffii strain ZS207 chromosome was carried out using Mauve74. For the phylogenetic reconstructions ClustalW and Neighbour-joining tree algorithm of MEGA7 was used with the default settings75.
AR determinants in WGS of M2a strain were searched using The Comprehensive Antibiotic Resistance Database (CARD)76, MEGARes77 and ARG-ANNOT78. INTEGRALL database79 and IS Finder80 were applied for searches of integrons and IS elements, respectively.
Completion of plasmid sequences and cloning of plasmids
For sealing the sequences of the putative plasmid scaffolds primers facing outward of the ends of scaffold sequences were designed (Suppl. Table S3) and PCRs were carried out using the respective primer pairs and plasmid DNA template isolated from M2a. PCR cycling was: initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 20 s, 55 °C for 30 s and 72 °C for 1 min and a final extension at 72 °C for 5 min. The PCR fragments obtained were sequenced on ABI Prism 3100 Genetic Analyzer (Perkin Elmer). The plasmid sequences assembled from scaffolds sc_93-98-170, sc_127-213-165, sc_167-103, and sc_219-191-168-115-278-239 could not be circularized even by long PCRs carried out using the appropriate primer pairs as follows: initial denaturation at 94 °C for 1.5 min followed by 10 cycles of 94 °C for 20 sec, 55 °C for 30 sec and 68 °C for 7 min, and 20 cycles of 94 °C for 20 sec, 55 °C for 30 sec and 68 °C for 7 min + 5 sec/cycle and a final extension at 68 °C for 10 min.
For cloning the circular plasmids into the R6K-based pir-dependent E. coli vector pSG76-C, a unique restriction site located out of the potential replication regions of each plasmid was applied. The plasmids pAVAci84 and pAVAci176 were linearized with PstI, pAVAci94 and pAVAci117 with SalI, pAVAci119 with XmaI and pAVAci145 with EcoRI and all were ligated into the corresponding sites of pSG76-C. pAVAci116 and pAVAci147 were cleaved with NruI and ligated into the SmaI site. pAVAci130 and pAVAci144 were cleaved with XhoI and MfeI, respectively, and ligated into the SalI and the EcoRI sites. The resulting pSG76-C derivative plasmids were maintained in E.coli TG2 λpir cells. The ability of cloned M2a-derived plasmids for autonomous replication in E. coli was tested by transformation into E. coli strain TG166. For selection of transformant E. coli strains 20 μg/ml chloramphenicol was used in the culture media.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files) or available in public databases.
References
Doughari, H. J., Ndakidemi, P. A., Human, I. S. & Benade, S. The Ecology, Biology and Pathogenesis of Acinetobacter spp.: An Overview. Microbes Environ. 26, 101–112 (2011).
Jung, J. & Park, W. Acinetobacter species as model microorganisms in environmental microbiology: current state and perspectives. Appl. Microbiol. Biotechnol. 99, 2533–2548 (2015).
Baumann, P., Doudoroff, M. & Stanier, R. Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J. Bacteriol. 95, 1520–1541 (1968).
Lessel, E. F. International Committee on Nomenclature of Bacteria Subcommittee on the Taxonomy of Moraxella and Allied Bacteria: Minutes of the Meeting, 11 August 1970. Room Constitution C, Maria-Isabel Hotel, Mexico City, Mexico. Int. J. Syst. Bacteriol. 21, 213–214 (1971).
Parte, A. C. & Road, W. LPSN — list of prokaryotic names with standing in nomenclature. 42, 613–616 (2014).
Juni, E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112, 917–931 (1972).
Janssen, P. et al. Discrimination of acinetobacter genomic species by AFLP fingerprinting. Int. J. Syst. Bacteriol. 47, 1179–1187 (1997).
Vaneechoutte, M. et al. Identification of Acinetobacter Genomic Species by Amplified Ribosomal DNA Restriction Analysis. J. Clin. Microbiol. 33, 11–15 (1995).
Dijkshoorn, L., van Harsselaar, B., Tjernberg, I., Bouvet, P. J. M. & Vaneechoutte, M. Evaluation of Amplified Ribosomal DNA Restriction Analysis for Identification of Acinetobacter Genomic Species. Syst. Appl. Microbiol. 21, 33–39 (1998).
Rafei, R. et al. Current molecular methods in epidemiological typing of Acinetobacter baumannii. Future Microbiol. 9, 1179–1194 (2014).
Bouvet, P. J. M. & Grimont, P. A. D. Taxonomy of the Genus Acinetobacter with the Recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and Emended Descriptions of Acinetobacter calcoaceticus a. Int. J. Syst. Bacteriol. 36, 228–240 (1986).
Nemec, A. et al. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans. Int. J. Syst. Evol. Microbiol. 59, 118–124 (2009).
Nemec, A. et al. Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int. J. Syst. Evol. Microbiol. 60, 896–903 (2010).
Nemec, A. et al. Revising the taxonomy of the Acinetobacter lwoffii group: The description of Acinetobacter pseudolwoffii sp. nov. and emended description of Acinetobacter lwoffii. Syst. Appl. Microbiol. 42, 159–167 (2019).
Atrouni, A. A., Joly-Guillou, M. L., Hamze, M. & Kempf, M. Reservoirs of non-baumannii Acinetobacter species. Front. Microbiol. 7, 1–12 (2016).
Carr, E. L., Kämpfer, P., Patel, B. K. C., Gürtler, V. & Seviour, R. J. Seven novel species of Acinetobacter isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53, 953–963 (2003).
Zarrilli, R., Pournaras, S., Giannouli, M. & Tsakris, A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11–19 (2013).
Antunes, L. C. S., Visca, P. & Towner, K. J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 71, 292–301 (2014).
Hamidian, M., Nigro, S. J., Hartstein, R. M. & Hall, R. M. RCH51, a multiply antibiotic-resistant Acinetobacter baumannii ST103IP isolate, carries resistance genes in three plasmids, including a novel potentially conjugative plasmid carrying oxa235 in transposon Tn6252. J. Antimicrob. Chemother. 72, 1907–1910 (2017).
Feng, G.-D., Yang, S.-Z., Wang, Y.-H., Deng, M.-R. & Zhu, H.-H. Acinetobacter guangdongensis sp. nov., isolated from abandoned lead-zinc ore. Int. J. Syst. Evol. Microbiol. 64, 3417–3421 (2014).
Mahjoubi, M. et al. Hydrocarbonoclastic bacteria isolated from petroleum contaminated sites In Tunisia: Isolation, identification and characterization of the biotechnological potential. N. Biotechnol. 30, 723–733 (2013).
Kostka, J. E. et al. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974 (2011).
Feng, G., Yang, S., Wang, Y., Yao, Q. & Zhu, H. Acinetobacter refrigeratorensis sp. nov., Isolated from a Domestic Refrigerator. Curr. Microbiol. 69, 888–893 (2014).
Gonzalez-Martinez, A. et al. Performance and microbial community structure of a polar Arctic Circle aerobic granular sludge system operating at low temperature. Bioresour. Technol. 256, 22–29 (2018).
Ventosa, A., Nieto, J. J. & Oren, A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62, 504–44 (1998).
Álvarez-Pérez, S., Lievens, B., Jacquemyn, H. & Herrera, C. M. Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov., isolated from floral nectar of wild Mediterranean insect-pollinated plants. Int. J. Syst. Evol. Microbiol. 63, 1532–1539 (2013).
Snowdon, J. A. & Cliver, D. O. Microorganisms in honey. Int. J. Food Microbiol. 31, 1–26 (1996).
Kim, P. S. et al. Acinetobacter apis sp. nov., isolated from the intestinal tract of a honey bee, Apis mellifera. J. Microbiol. 52, 639–645 (2014).
Metzgar, D. et al. Acinetobacter sp. ADP1: An ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32, 5780–5790 (2004).
Jacobs, A. C., Thompson, M. G. & Black, C. C. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a. MBio 5, 1–10 (2014).
Abdel-el-haleem, D. Minireview Acinetobacter: environmental and biotechnological applications. African. J. Biotechnol. 2, 71–74 (2003).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–32 (2005).
Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304 (2013).
Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 31, e00088–17 (2018).
Roca, I., Espinal, P., Vila-Fanés, X. & Vila, J. The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3, 1–30 (2012).
Nowak, P. & Paluchowska, P. Acinetobacter baumannii: Biology and drug resistance — role of carbapenemases. Folia Histochem. Cytobiol. 54, 61–74 (2016).
Lean, S. S. & Yeo, C. C. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: Good, bad, who knows? Front. Microbiol. 8, 1–8 (2017).
Seifert, H., Schulze, A., Baginski, R. & Pulverer, G. Plasmid DNA fingerprinting of Acinetobacter species other than Acinetobacter baumannii. J. Clin. Microbiol. 32, 82–86 (1994).
Gerner-Smidt, P. Frequency of plasmids in strains of Acinetobacter calcoaceticus. J. Hosp. Infect. 14, 23–28 (1989).
Nigro, S. J. & Hall, R. M. Structure and Context of acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 71, 1135–1147 (2016).
Zou, D. et al. Complete sequences of two novel bla NDM-1-harbouring plasmids from two Acinetobacter towneri isolates in China associated with the acquisition of Tn125. Sci. Rep. 7, 9405 (2017).
Koczura, R., Przyszlakowska, B., Mokracka, J. & Kaznowski, A. Class 1 integrons and antibiotic resistance of clinical Acinetobacter calcoaceticus-baumannii complex in Poznań, Poland. Curr. Microbiol. 69, 258–262 (2014).
Hamidian, M., Holt, K. E. & Hall, R. M. Genomic resistance island AGI1 carrying a complex class 1 integron in a multiply antibiotic-resistant ST25 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 70, 2519–2523 (2015).
Kurakov, A. et al. The ancient small mobilizable plasmid pALWED1.8 harboring a new variant of the non-cassette streptomycin/spectinomycin resistance gene aadA27. Plasmid 84–85, 36–43 (2016).
Veress, A., Wilk, T., Kiss, J., Olasz, F. & Papp, P. P. Draft Genome Sequences of Saccharibacter sp. Strains 3.A.1 and M18 Isolated from Honey and a Honey Bee (Apis mellifera) Stomach. Genome Announc. 5 (2017).
Veress, A., Wilk, T., Kiss, J., Papp, P. P. & Olasz, F. Two Draft Genome Sequences of Sphingobacterium sp. Strains Isolated from Honey. Genome Announc. 5, 4–5 (2017).
Turton, J. F., Hyde, R., Martin, K. & Shah, J. Genes encoding OXA-134-like enzymes are found in Acinetobacter lwoffii and A. schindleri and can be used for identification. J. Clin. Microbiol. 50, 1019–1022 (2012).
Figueiredo, S. et al. OXA-134, a naturally occurring carbapenem-hydrolyzing class D β-lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 54, 5372–5375 (2010).
Dillon, B., Thomas, L., Mohmand, G., Zelynski, A. & Iredell, J. Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 62, 221–232 (2005).
Koeleman, J. G. M., Stoof, J., Van Der Bijl, M. W., Vandenbroucke-Grauls, C. M. J. E. & Savelkoul, P. H. M. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 39, 8–13 (2001).
Vallenet, D. et al. Comparative analysis of acinetobacters: Three genomes for three lifestyles. PLoS One 3 (2008).
Mindlin, S. et al. Resistance of Permafrost and Modern Acinetobacter lwoffii Strains to Heavy Metals and Arsenic Revealed by Genome Analysis. Biomed Res. Int. 2016 (2016).
Thomas, C. M. Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491 (2000).
Li, R., Chen, K., Wai, E., Chan, C. & Chen, S. Resolution of dynamic MDR structures among the plasmidome of Salmonella using MinION single-molecule, long-read sequencing. 10–14, https://doi.org/10.1093/jac/dky243 (2018).
Kiss, J. et al. Identification and Characterization of oriT and Two Mobilization Genes Required for Conjugative Transfer of Salmonella Genomic Island 1. Front. Microbiol. 10, 1–16 (2019).
Heuer, H., Kopmann, C., Binh, C. T. T., Top, E. M. & Smalla, K. Spreading antibiotic resistance through spread manure: Characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 11, 937–949 (2009).
Heuer, H. & Smalla, K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36, 1083–1104 (2012).
Kyselková, M. et al. Characterization of tet(Y)-carrying lowGC plasmids exogenously captured from cow manure at a conventional dairy farm. FEMS Microbiol. Ecol. 92, 1–10 (2016).
Hamidian, M., Nigro, S. J. & Hall, R. M. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 67, 2833–2836 (2012).
Regalado, N. G., Martin, G. & Antony, S. J. Acinetobacter lwoffii: Bacteremia associated with acute gastroenteritis. Travel Med. Infect. Dis. 7, 316–317 (2009).
Zou, D. et al. A Novel New Delhi Metallo-β-Lactamase Variant, NDM-14, Isolated in a Chinese Hospital Possesses Increased Enzymatic Activity against Carbapenems. Antimicrob. Agents Chemother. 59, 2450–2453 (2015).
Mittal, S., Sharma, M., Yadav, A., Bala, K. & Chaudhary, U. Acinetobacter lwoffii An Emerging Pathogen in Neonatal ICU. Infect. Disord. - Drug Targets 15, 184–188 (2015).
De Man, J. C., Rogosa, M. & Sharpe, M. E. A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 23, 130–135 (1960).
Ito, A. et al. In Vitro Antibacterial Activity of AM-715, a New Nalidixic Acid Analog. Antimicrob. Agents Chemother. 17, 103–108 (1980).
Vashist, H., Sharma, D. & Gupta, A. A review on commonly used biochemical test for bacteria. Innovare J. Life Sci. 1, 1–7 (2013).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. New York (1989).
Pósfai, G., Koob, M. D., Kirkpatrick, H. A. & Blattner, F. R. Versatile insertion plasmids for targeted genome manipulations in bacteria: Isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179, 4426–4428 (1997).
Simon, R., Priefer, U. & Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1, 784–791 (1983).
Kiss, J., Nagy, B. & Olasz, F. Stability, entrapment and variant formation of Salmonella genomic island 1. PLoS One 7, e32497 (2012).
Lane, D. J. 16S/23S rRNA sequencing, p. 115–175. In Stackebrandt, E. & Goodfellow, M. (eds.) Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons, Chichester, New York. (1991).
Coil, D., Jospin, G. & Darling, A. E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–9 (2015).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–10 (1990).
Darling, A. C. E. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 14, 1394–1403 (2004).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–4 (2016).
McArthur, A. G. et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–57 (2013).
Lakin, S. M. et al. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 45, D574–D580 (2017).
Gupta, S. K. et al. ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220 (2014).
Moura, A. et al. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–8 (2009).
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J. & Chandler, M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–6 (2006).
Acknowledgements
We thank Erika Sztánáné-Keresztúri for the excellent technical assistance and Tibor Farkas and Agatha Treveil for their comments and proofreading the text. This work was supported by the Hungarian Scientific Research Fund K 105635 and NKFI K 128203 to J. Kiss. A.V. and T.W. were supported by the PhD fellowship of Eötvös Loránd University, Budapest, Hungary.
Author information
Authors and Affiliations
Contributions
F.O. conceived the project and managed the WG sequencing. J. Kiss and A.V. designed the experiments and analyzed the data. A.V. and J. Kömüves carried out the experiments. T.N. and T.W. performed the bioinformatics. J. Kiss wrote the paper and prepared the figures and all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Veress, A., Nagy, T., Wilk, T. et al. Abundance of mobile genetic elements in an Acinetobacter lwoffii strain isolated from Transylvanian honey sample. Sci Rep 10, 2969 (2020). https://doi.org/10.1038/s41598-020-59938-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59938-9
This article is cited by
-
Complex secondary structure in small Rep_3 plasmids of Acinetobacter spp.
Biologia (2023)
-
Remarkable shift in structural and functional properties of an animal charcoal-polluted soil accentuated by inorganic nutrient amendment
Journal of Genetic Engineering and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.