Abstract
The primary hurdle for diagnosis of some diseases is the long incubation required to culture and confirm the presence of bacteria. The concept of using microbial VOCs as “signature markers” could provide a faster and noninvasive diagnosis. Finding biomarkers is challenging due to the specificity required in complex matrices. The objectives of this study were to (1) build/test a lab-scale platform for screening of microbial VOCs and (2) apply it to Mycobacterium avium paratuberculosis; the vaccine strain of M. bovis Bacillus Calmette-Guérin; and M. kansasii to demonstrate detection times greater those typically required for culture. SPME-GC-MS was used for sampling, sample preparation, and analyses. For objective (1), a testing platform was built for headspace sampling of bacterial cultures grown in standard culture flasks via a biosecure closed-loop circulating airflow system. For (2), results show that the suites of VOCs produced by Mycobacteria ssp. change over time and that individual strains produce different VOCs. The developed method was successful in discriminating between strains using a pooled multi-group analysis, and in timepoint-specific multi- and pair-wise comparisons. The developed testing platform can be useful for minimally invasive and biosecure collection of biomarkers associated with human, wildlife and livestock diseases for development of diagnostic point-of-care and field surveillance.
Similar content being viewed by others
Introduction
Bovine tuberculosis (bTB) is a zoonotic disease of international public health, trade, agricultural, and wildlife management significance1,2. The disease is caused by Mycobacterium bovis, a member of the Mycobacterium tuberculosis complex3,4,5. It is estimated that in 2014, 9.6 million new cases human tuberculosis (hTB) and 1.5 million associated deaths occurred worldwide6, with the majority caused by M. tuberculosis; however, zoonotic tuberculosis is often under-reported. Muller et al. 2013 reported that approximately 10–37% of hTB cases may be caused by M. bovis infection, especially in developing countries where the prevalence of livestock bTB may reach 10–14%7,8,9,10,11,12.
A primary hurdle for mycobacterial disease diagnosis is the long incubation time required to culture and confirm the presence of mycobacteria in biological samples. Culture may take approximately eight weeks before final results can be confirmed. Several in vitro blood based tests (i.e., interferon-ɣ release assay) have been developed to confirm that an individual has been exposed to mycobacteria. However, these tests suffer from cross-reactivity with closely related non-tuberculous mycobacteria resulting in false positive test results. Development of technologies to reduce the time between the start of culture, detecting growth, and positively identifying the mycobacterial agent will improve both the diagnosis and appropriate treatment of infected humans, and eradication of bTB from wildlife and livestock.
In recent years increased attention has been given to the concept of using microbial volatile organic compound (VOC) emissions as “signature markers” (a.k.a. biomarkers) for faster, more economical, and noninvasive disease diagnosis in humans and animals. These VOC emissions may be collected from breath, blood, skin, urine, feces and other bodily secretions. Studies have identified potential VOC biomarkers related to multiple diseases such as cholera, cancer, diabetes, uremia, schizophrenia, asthma, liver disease, chronic lung disease, Pseudomoniasis, tuberculosis, and others13,14,15,16. Several methods of collecting and analyzing VOCs for potential diagnosis of M. tuberculosis and M. bovis have been described. Closed loop stripping analysis (CLSA) - gas chromatography- mass spectrometry (GC-MS) was used to detect VOCs from multiple strains of M. tuberculosis from cultures17, and select ion flow tube (SIFT)-MS has been used to measure VOCs present in the headspace of M. bovis BCG cultures18 and to measure VOCs in the breath of children with cystic fibrosis14, NH3 levels in human breath for Helicobacter pylori screening, and detect acetonitrile levels in smokers’ breath19.
Electronic ‘e-nose’ technology has been used for detection of VOCs present in sputum samples collected from humans infected with M. tuberculosis14, and VOCs from the headspace of cultures of M. bovis BCG and M. smegmatis18. ‘E-nose’ has also been reported capable of differentiating between M. tuberculosis, three other bacteria, and a control19, monitoring smokers’ habits by measuring breath CO, detecting H. pylori presence in association with chronic gastritis, and detecting N2O produced by respiratory inflammation16. Thermal desorption (TD)-GC-MS has been used to determine VOCs present in the headspace of M. bovis BCG cultures18, and cattle breath20; and solid-phase microextration (SPME)-GC-MS was used to determine biomarkers M. tuberculosis and M. bovis in cultures and M. tuberculosis in human breath21,22. Other methods that have been used to collect VOCs associated with diseases that could be utilized in the future for TB detection include ion mobility spectrometry (IMS)14,23, proton transfer reaction (PTR)-MS14,15,16, and laser spectroscopy24. Select ion flow tube-MS, ‘e-noses’, IMS, PTR-MS have the advantage of being fast and potentially mobile. The downside of these methods includes decreased sensitivity, and the inability to chemically identify or profile VOCs. GC-MS-based methods may be slower, more expensive, and the instrumentation is not typically mobile; however, they have the advantage of being able to reproducibly identify and profile known and unknown microbial VOCs at low concentration ranges14,16.
Solid-phase microextration is an attractive technology for collecting microbial VOCs due to its simplicity, ease of use, and ability to sample and pre-concentrate a wide range of potential target compounds. Dynamic headspace extraction of VOCs can increase mass transfer to the SPME fiber compared to static headspace extraction, thus, reducing sampling times while improving mass loading of the fiber25,26. In contrast, both CLSA and TD are capable of pre-concentrating VOCs but require extra equipment and are more labor intensive than SPME. Syhre et al.21 collected VOCs from seven mycobacterial and 16 other respiratory pathogen cultures using three different SPME fiber types; 100 µm polydimethylsiloxane (PDMS), 2 cm 50/30 µm divinylbenzene (DVB)/Carboxen/PDMS, and 70 µm Carbowax/DVB. The SPME fiber coated with 2 cm 50/30 µm DVB/Carboxen/PDMS was found to recover higher concentrations of all target VOCs. Other studies have utilized the 50/30 µm DVB/Carboxen/PDMS-type and Carboxen/PDMS-type SPME fiber coatings for microbial VOC collection in human and cattle breath samples22,27,28, in bovine nasals excretions29, and in bovine fecal excretions from cattle vaccinated with M. bovis challenge30. SPME has been also used for the in-vivo and in-vitro collection of rumen gases31,32, VOCs emitted from wildlife marking fluids33,34, decaying carcasses35.
The objectives of this research were to (1) design, build, and test a lab-scale dynamic VOC sampling platform specifically capable of simultaneous biosecure SPME collection of headspace VOCs emitted from controlled bacterial cultures and a media control; (2) apply this screening method to M. avium paratuberculosis (MAP); the vaccine strain of M. bovis Bacillus Calmette-Guérin (BCG); and M. kansasii cultures to demonstrate a proof-of-concept detection method that is faster than standard culture methods.
Our first working hypothesis was that by sampling the recirculating culture headspace air, we would be able to detect trace-levels of microbial VOCs early in the incubation process with minimal background interference. Our second hypothesis was that SPME sampling of microbial VOCs followed by GC-MS analysis would be suitable to detect differences in microbial VOCs emitted by different cultured strains of mycobacteria.
If successful, this proof-of-concept identification of VOC biomarkers would allow differentiation between the microbial agents prior to the eight weeks often required for cultured mycobacterial strain identification. The knowledge gained from this work could be directly applied for diagnosis of hTB and bTB, for detection and identification of VOC biomarkers produced by culture of other pathogenic bacteria, and as a reference library for pathogen-produced VOCs present in other samples such as breath, feces, urine, blood, and other biofluids.
Results and Discussion
Reducing interfering background VOCs in lab-scale testing platform
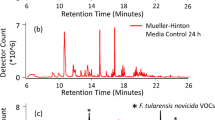
To minimize the VOC background and improve the likelihood of finding unique microbial VOCs for Objective 2 the prototype sampling platform was exposed to an initial 21 h bake-out at 50 °C, which only decreased the background slightly. After bake-out of the Neoprene tubing, system background emissions were reduced by 75% (Fig. 1). Background emissions present in the microbial growth media were also determined as reproducible and necessary background populated by numerous VOCs with relatively low peak areas.
Multi-group analysis of single ion data
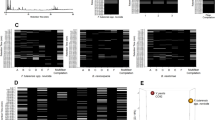
A total of 123 fragmentation ions were identified by XCMS Online, with 33 meeting the criteria for statistical significance (α = 0.05; fold change ≥1.5). Visualization of the statistically significant ion feature characteristics are depicted by XCMS Online as a cloud plot (Fig. 2)36. Briefly, all sample chromatograms were aligned and overlaid onto the x-axis. Significant ion features are identified as circles, with ions with greatest m/z ratios located furthest from the x-axis. Circle size is proportional to the degree of fold change (larger circle = greater fold change), while color intensity corresponds to the statistical significance (p-value) of the fold change as calculated by a Welch t-test with unequal variances (darker color = lower p-value).
Identification of culture specific peak area data
Statistically significant ion intensities were matched by retention time to 77 total ion chromatographic (TIC) peaks. For each culture, trends in the changes of potential VOCs TIC intensities across the three time-points in each of the replicates were graphed. This allowed identification of ten VOC compounds with consistent, repeatable changes across time in each of the three replicates (3 studies) (Figs 3, 4 and 5).
Weekly trends of three compounds found in M. bovis Bacillus Calmette-Guérin (BCG) cultures. Note: (A): Compound 1 identified at retention time 4.250 min, (B): Compound 2 identified at retention time 9.286 min, (C): Compound 3 identified at retention time 17.080 min. Black solid line: Replicate 1, Dotted black line: Replicate 2, Gray solid line: Replicate 3.
Weekly trends of two compounds identified in Mycobacterium avium paratuberculosis cultures. Note: (A): Compound 1 identified at retention time 5.309 min, (B): Compound 2 identified at retention time 9.286 min. Solid black line: Replicate 1, Dotted black line: Replicate 2, Solid gray line: Replicate 3.
Weekly trends of five compounds identified in M. kansasii cultures. Note: (A): Compound 1 identified at retention time 5.309 min, (B): Compound 2 identified at retention time 11.344 min, (C): Compound 3 identified at retention time 18.769 min, (D): Compound 4 identified at retention time 19.048 min, (E): Compound 5 identified at retention time 20.114 min. Black solid line: Replicate 1, Dotted black line: Replicate 2, Solid gray line: Replicate 3.
Tentative identification of biomarkers
Tentative identifications of peaks and associated information are summarized in Table 1. Tentatively identified compounds include two aldehydes (octanal, decanal); two alkanes (3,3-dimethyl hexane, tetradecane); three benzenes (benzene, ethylbenzene, styrene); one dicarboxylic acid (pentanedioic acid, 2,4-dimethyl dimethyl ester); one isothiocyanate (cyclohexane, isothiocyanate); two ketones (2-pentanone, acetophenone); and one oxotane (lilac aldehyde B). Two unique (benzene, acetophenone) and one shared (ethylbenzene; with MAP) are identified in BCG cultures. Two shared compounds (2-pentanone, M. kansasii; ethylbenzene, BCG) are associated with MAP cultures, while four unique (styrene; pentanedioic acid, 2, 4-dimethyl, dimethyl ester; 3,3-dimethyl hexane; cyclohexane, isothiocyanate) and one shared compound (2-pentanone, MAP) are associated with M. kansasii cultures. Four unique compounds (octanal, decanal, tetradecane, lilac aldehyde D) appear in the control media.
The four compounds unique to the control media were removed from further evaluation, leaving ten mycobacterial-associated compounds to evaluate for potential bacterial metabolic or physiologic associations (Table 2).
There has been little research exploring the metabolic pathways of M. bovis BCG, MAP, and M. kansasii, therefore, published literature exploring the metabolome of M. tuberculosis complex and environmental strains of mycobacteria were considered sources for comparison (Table 3).
Conclusions
This study demonstrates a proof-of-concept for the detection and use of microbial VOCs as a means to discriminate between mycobacterial cultures associated with one to three week post-culture inoculation, a time span preceding the time required to currently identify pathogenic M. tuberculosis and M. bovis in diagnostic cultures. To accomplish this task (Objective 1), a sampling system was designed, built, and tested for controlled collection of microbial volatiles using closed-loop headspace airflow over microbial cultures, sampling and sample preparation with SPME, analyte separation and identification via GC-MS, and tentative compound identification using novel metabolomics databases and the NIST W8N08 library. The capability of this system to produce results more efficiently than some currently utilized diagnostic modalities such as culture exemplifies its potential use as a diagnostic tool. The lab-scale testing platform concept can be useful for minimally invasive and biosecure collection of marker volatiles associated with human, wildlife and production animal diseases for development of diagnostic non-invasive point-of-care tools, field surveillance technologies and strategies.
Objective 2 provided a comprehensive assessment of VOCs collected from the headspace of three different mycobacterial cultures and one control media sample. Discrimination between mycobacterial cultures was successful one, two, and three weeks post-culture inoculation, a time span preceding the time required to currently identify pathogenic M. tuberculosis and M. bovis in diagnostic cultures. Unique VOCs representing potential biomarkers were identified in two mycobacterial cultures (e.g., BCG, M. kansasii). No unique plausible biomarkers were identified for the MAP cultures; however, discrimination from the two other mycobacterial cultures was possible when the VOC profiles of all the cultures were examined in context.
From a diagnostic perspective, detection of VOCs produced by pathogenic mycobacteria at early states of culture growth could improve disease diagnosis and treatment, especially in developing countries where access to sophisticated laboratory diagnostics is limited. The capability to differentiate between human and zoonotic mycobacteria under such circumstances could improve the capability of physicians to more accurately diagnose tuberculosis patients, to differentiate between hTB and zoonotic bTB, and to appropriately dispense medication targeted toward the etiological disease agent.
Materials and Methods
Experimental design
The experimental part of this study was carried out at the USDA-ARS National Animal Disease Center (NADC) and the Atmospheric Air Quality Laboratory of Iowa State University (ISU) in accordance with the Guide for the Institutional Animal Care and Use Committee. The protocol was approved by Iowa State University’s Institutional Animal Care and Use Committee (IACUC Log # 4-14-7787-B) and Institutional Biosafety Committee (ID: 14-I-015-A/H).
The proof-of-concept study was designed to determine variations and identify specific VOCs produced by growing mycobacterial cultures. Mycobacterial strains included M. avium paratuberculosis (MAP; Strain K10); M. bovis Bacillus Calmette-Guérin (BCG) (Danish 1331); and M. kansasii (Strain 03-6931). Approximately one optical density of each mycobacterial strain was added to respective 225 mL (culture surface area, 75 cm2) culture bottles (430725, Corning®, Corning, New York, USA) containing 30 mL of Middlebrook 7H9 media enriched with 10% Middlebrook OADC, 0.05% Tween-80 and 2 mg mL−1 of Mycobactin J. Cultures were incubated at 37 °C on the testing platform in an incubator for three weeks. Dynamic headspace samples of each culture bottle were collected weekly with SPME. A control consisting of only growth media was used to determine “background” VOCs.
Lab-scale testing platform
The lab-scale testing platform was designed, built and tested using several general lab use components. The reusable testing platform consisted of four Omegaflex FPU100 peristaltic pumps (Omega Engineering Inc., Stamford, CT, USA) to circulate air (average flow rate of 162 mL min−1) through each modified culture bottle in closed loops with each separate loop containing an inline ~13 mL, glass sampling bulb (28526-U, Supelco, Bellefonte, PA, USA) for SPME extraction (Fig. 6). Each closed loop was constructed from PTFE Teflon tubing from the pumps to the culture bottles, to the sampling bulbs, back to the pumps; and Neoprene tubing sections in the peristaltic pumps. The inlet and outlet fittings on each culture bottle included 0.22 micron filters to prevent introduction of mycobacteria into the closed loop system and rendering the system to be self-contained and biosecure. The platform’s compact size enabled the entire system to be placed in an incubator for optimal temperature for growing mycobacteria. The materials used to constuct the platform allowed relatively easy decontamination after and between-the-trials use.
Microbial volatile organic compounds
A 2 cm 50/30 µm DVB/Carboxen/PDMS fiber (57348-U, Supelco, Bellefonte, PA, USA) SPME fiber was used to extract and pre-concentrate VOCs from circulating gas. Samples were collected by dynamic headspace extraction with SPME for 1 h at 37 °C using the lab-scale testing platform. After the sampling period, the SPME fiber with extracted VOCs was inserted into the 250 °C GC injector for 2 min for thermal desorption, sample introduction, analyte separation and analysis.
The multidimensional GC–MS-Olfactometry (MDGC-MS-O) system (Microanalytics, Volatile Analysis Corporation, Round Rock, TX, USA) used for analysis was equipped with two columns connected in series. The non-polar pre-column was 30 m × 0.53 mm i.d.; film thickness, 0.50 µm with 5% phenyl polysilphenylene siloxane stationary phase (SGE BPX-5) and operated with constant pressure mode at 13.5 psi (0.92 atm). The polar analytical column was 30 m × 0.53 mm bonded polyethylene glycol (PEG) embedded in a synthetic glass (SGE SolGel-Wax) at a film thickness of 0.50 µm. System automation and data acquisition software were MultiTraxTM V. 10.1 (Microanalytics, Volatile Analysis Corporation, Round Rock, TX, USA) and ChemStation™ (Agilent Technologies, Santa Clara, CA, USA). The GC run parameters were as follows: injector, 250 °C; column, 40 °C initial, 3 min hold, 7 °C min−1 ramp to 240 °C final, 8.43 min hold; carrier gas, UHP-grade helium (99.999%). The GC was operated in a constant pressure mode where the mid-point pressure, i.e., pressure between pre-column and analytical column, was always at 5.7 psi (0.39 atm) and the heart-cut sweep pressure was 5.0 psi. This type of columns configuration (in series with different polarity) does not lend itself to a classic retention index (RI) approach for tentative compound identification with n-alkanes. The MS full scan range was 34 to 150 m z−1. Spectra were collected at 2 scans s−1 using full scan. The quadrupole MS was set to electron ionization (EI) mode with ionization energy of 70 eV. MS tuning was performed using the default autotune setting using perfluorotributylamine (PFTBA) daily.
Reducing interfering background VOCs in lab-scale testing platform
After construction, the platform’s background VOCs were initially baked-out for 21 h at 50 °C (the maximum temperature that some plastic components in the platform could withstand) The Neoprene tubing in the peristaltic pumps was removed from the platform and baked-out separately at 110 °C for 18 h to remove any VOCs the might be released into the closed platform loop.
Data analysis
Cultures were identified in raw data as culture 1–4 and sample collection time-point (Week 1, Week 2, Week 3) until the data analysis was complete to prevent bias. Total ion chromatograms (TICs) from all cultures at all time-points were analyzed using the multi-group comparison feature in XCMS Online to identify peak ion abundances that differed between the cultures at each time-point37. Statistically significant ion intensities were matched to GC column retention time chromatographic peaks using Agilent Mass Hunter software (Agilent Technologies, Santa Clara, CA, USA). Peak areas were determined using the TICs. Peak area data for each culture at each replicate and time-point were evaluated to determine the suite of VOCs best suited to provide optimal discrimination in multi-group and pair-wise comparisons. Peaks were tentatively identified using AMDIS deconvolution software38, the National Institute of Standards and Technology (NIST) W8N08 database39, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database40,41,42, and the Human Metabolome Database (HMDB)43,44,45,46. Tentative compound identifications were made based on ≥65% match with compounds present in these libraries. Tentative metabolic sources for each compound were explored using KEGG, HMDB, and review of peer-reviewed literature47.
References
Biet, F., Boschiroli, M. L., Thorel, M. F. & Guilloteau, L. A. Zoonotic aspects of Mycobacterium bovis and Mycobacterium avium-intracellulare complex (MAC). Vet. Res. 36, 411–436 (2005).
Schiller, I. et al. Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound. Emerg. Dis. 57, 205–220 (2010).
Cole, S. T. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology. 148, 2919–2928 (2002).
Mostowy, S., Cousins, D., Brinkman, J., Aranaz, A. & Behr, M. A. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186, 74–80 (2002).
Brosch, R. et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. 99, 3684–3689 (2002).
Global Tuberculosis Report, World Health Organization (2015).
Müller, B. et al. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg. Infect. Dis. 19, 899–908 (2013).
Cosivi, O. et al. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4, 59–70 (1998).
Cleaveland, S. et al. Mycobacterium bovis in rural Tanzania: Risk factors for infection in human and cattle populations. Tuberculosis. 87, 30–43 (2007).
de la Rua-Domenech, R. et al. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 81, 190–210 (2006).
Grange, J. M. Mycobacterium bovis infection in human beings. Tuberculosis. 81, 71–77 (2001).
Michel, A. L., Müller, B. & Van Helden, P. D. Mycobacterium bovis at the animal–human interface: A problem, or not? Vet. Microbiol. 140, 371–381 (2010).
Shirasu, M. & Touhara, K. The scent of disease: volatile organic compounds of the human body related to disease and disorder. J. Biochem. 150, 257–266 (2011).
Sethi, S., Nanda, R. & Chakraborty, T. Clinical application of volatile organic compound analysis for detecting infectious diseases. Clin. Microbiol. Rev. 26, 462–475 (2013).
Thorn, R. M. S. & Greenman, J. Microbial volatile compounds in health and disease conditions. J. Breath Res. 6, 024001 (2012).
Krisher, S., Riley, A. & Mehta, K. Designing breathalyser technology for the developing world: how a single breath can fight the double disease burden. J. Med. Eng. Technol. 38, 156–163 (2014).
Nawrath, T., Mgode, G. F., Weetjens, B., Kaufmann, S. H. & Schulz, S. The volatiles of pathogenic and nonpathogenic mycobacteria and related bacteria. Beilstein J. Org. Chem. 8, 290–299 (2012).
McNerney, R., Mallard, K., Okolo, P. I. & Turner, C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol. Lett. 328, 150–156 (2012).
Pavlou, A. K. et al. Detection of Mycobacterium tuberculosis (TB) in vitro and in situ using an electronic nose in combination with a neural network system. Biosens. Bioelectron. 20, 538–544 (2004).
Ellis, C. K. et al. A pilot study exploring the use of breath analysis to differentiate healthy cattle from cattle experimentally infected with Mycobacterium bovis. PLoS One. 9, e89280 (2014).
Syhre, M. & Chambers, S. T. The scent of Mycobacterium tuberculosis. Tuberculosis. 88, 317–323 (2008).
Syhre, M., Manning, L., Phuanukoonnon, S., Harino, P. & Chambers, S. T. The scent of Mycobacterium tuberculosis–part II breath. Tuberculosis. 89,, 263–266 (2009).
Purkhart, R. et al. Chronic intestinal Mycobacteria infection: discrimination via VOC analysis in exhaled breath and headspace of feces using differential ion mobility spectrometry. J. Breath Res. 5, 027103 (2011).
Kim, K.-H., Jahan, S. A. & Kabir, E. A review of breath analysis for diagnosis of human health. Trends Anal. Chem. 33, 1–8 (2012).
Augusto, F., Koziel, J. A. & Pawliszyn, J. Design and validation of portable SPME devices for rapid field air sampling and diffusion based calibration. Anal. Chem. 73, 481–486 (2001).
Pawliszyn, J. (Ed.), Handbook of solid phase microextraction. (Chemical Industry Press, 2009).
Spinhirne, J. P., Koziel, J. A. & Chirase, N. K. Sampling and analysis of volatile organic compounds in bovine breath by solid-phase microextraction and gas chromatography–mass spectrometry. J. Chromatogr. A. 1025, 63–69 (2004).
Spinhirne, J. P., Koziel, J. A. & Chirase, N. K. A device for noninvasive on-site sampling of cattle breath with solid phase microextraction. Biosystems Engineering. 84, 239–246 (2003).
Maurer, D. L., Koziel, J. A., Engelken, T. J., Cooper, V. L. & Funk, J. L. Detection of volatile compounds emitted from nasal secretions and serum: towards non-invasive identification of diseased cattle biomarkers. Separations. 5, 18 (2018).
Ellis, C. K. et al. Use of fecal volatile organic compound analysis to discriminate between non-vaccinated and BCG-vaccinated cattle prior to and after Mycobacterium bovis challenge. PLoS One 12, e0179914 (2017).
Cai, L. et al. Characterization of VOCs and odors by in vivo sampling of beef cattle rumen gas using SPME and GC-MS-olfactometry. Anal. Bioanal. Chem. 386, 1791–1802 (2006).
Spinhirne, J. P., Koziel, J. A. & Chirase, N. Characterizing volatile fatty acids and other gases in a rumen closed in vitro fermentation system using solid phase microextraction. Trans. ASABE. 46, 585–588 (2003).
Soso, S. B. & Koziel, J. A. Analysis of odorants in marking fluid of Siberian tiger (Panthera tigris altaica) using simultaneous sensory and chemical analysis with headspace solid-phase microextraction and multidimensional gas chromatography-mass spectrometry-olfactometry. Molecules. 21, 834 (2016).
Soso, S. B. & Koziel, J. A. Characterizing the scent and chemical composition of Panthera leo marking fluid using solid-phase microextraction and multidimensional gas chromatography-mass spectrometry-olfactometry. Sci. Rep. 7, 5137 (2017).
Koziel, J. A. et al. Method for sampling and analysis of volatile biomarkers in process gas from aerobic digestion of poultry carcass using time-weighted average SPME and GC-MS. Food Chem. 232, 799–807 (2017).
Patti, G. J. et al. A view from above: cloud plots to visualize global metabolomic data. Anal. Chem. 85, 798–804 (2012).
The Scripps Research Institute. XCMS. Retrieved from, https://xcmsonline.scripps.edu (2016, March).
AMDIS. Retrieved from, http://www.amdis.net (2018, December 27).
U.S. Department of Commerce. National Institute of Standards and Technology. Retrieved from, http://www.nist.gov (2016, July 18).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa Laboratories. Kyoto Encyclopedia of Genes and Genomes. Retrieved from, http://www.genome.jp/kegg/ (2016, July 1).
Aoki, K. F., Kanehisa, M. Using the KEGG database resource. Current Protocols in Bioinformatics. (John Wiley & Sons, Inc., 2002).
The Metabolomics Innovation Center. Human Metabolomics Data Base. Retrieved from, http://www.hmdb.ca (2016).
Wishart, D. S. et al. HMDB: The human metabolome database. Nucleic Acids Res. 35, D521–D526 (2007).
Wishart, D. S. et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 37, D603–D610 (2009).
Wishart, D. S. et al. HMDB 3.0-the human metabolome database in 2013. Nucleic Acids Res. 41, D801–D807 (2013).
SRI International. Biocyc Database Collection. Retrieved from, http://biocyc.org/ (2016).
Trefz, P. et al. Volatile emissions from Mycobacterium avium subsp. paratuberculosis mirror bacterial growth and enable distinction of different strains. PLoS One. 8, e76868 (2013).
Bergmann, A. et al. In Vivo volatile organic compound signatures of Mycobacterium avium subsp. paratuberculosis. PLoS One. 10, e0123980 (2015).
Fischer, S. et al. Physiological variability in volatile organic compounds (VOCs) in exhaled breath and released from faeces due to nutrition and somatic growth in a standardized caprine animal model. J. Breath Res. 9, 027108 (2015).
Zhang, L. et al. Biodegradation of benzene, toluene, ethylbenzene, and o-xylazene by the bacterium Mycobacterium cosmeticum byf-4. Chemosphere. 90, 1340–1347 (2013).
Burback, B. & Perry, J. J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. J Appl. Environ. Microbiol. 59, 1025–1029 (1993).
Phillips, M. et al. Volatile biomarkers of pulmonary tuberculosis in the breath. Tuberculosis. 87, 44–52 (2007).
Chownk, M. et al. mesT, a unique epoxide hydrolase is essential for optimal growth of Mycobacterium tuberculosis in the presence of styrene oxide. Future Microbiology. 12, 527–546 (2017).
Wilkin, J. M. et al. Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase. Eur. J. Biochem. 262, 299–307 (1999).
Galamba, A. et al. Molecular and biochemical characterization of Mycobacterium smegmatis alcohol dehydrogenase C. FEMS Microbiology Letters 196, 51–56 (2001).
Stahl, R. S. et al. Fecal volatile organic compound profiles from white-tailed deer (Odocoileus virginianus) as indicators of Mycobacterium bovis exposure or Mycobacterium bovis Bacille Calmette-Guerin (BCG) vaccination. PLoS One. 10, e0129740 (2015).
Agerbirk, N. & Olsen, C. E. Glucosinolate structures in evolution. Phytochemistry. 77, 16–45 (2012).
Phillips, M. et al. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis. 90, 145–151 (2010).
Phillips, M. et al. Point-of-care breath test for biomarkers of active pulmonary tuberculosis. Tuberculosis. 92, 314–320 (2012).
Banday, K. M. et al. Use of urine volatile organic compounds to discriminate tuberculosis patients from healthy subjects. Anal. Chem. 83, 5526–5534 (2011).
Crespo, E. et al. Potential biomarkers for identification of mycobacterial cultures by proton transfer reaction mass spectrometry analysis. Rapid Commun. Mass Spectrom. 26, 679–685 (2012).
Kolk, A. H. J. et al. Breath analysis as a potential diagnostic tool for tuberculosis. Int. J. Tuberc. Lung Dis. 16, 777–782 (2012).
Acknowledgements
This research was partially funded by the USDA-APHIS-sponsored project # 15-7488-1156-CA ‘Development of GC/MS method to detect disease-related volatile organic compounds in breath and manure samples of cattle and white-tailed deer’ (2015–2016).
Author information
Authors and Affiliations
Contributions
Devin L. Maurer conceived and built the lab-scale testing platform as well as preparation, running and analysis of samples, Christine K. Ellis conceived the research and carried out the analysis and identification of GC-MS identified compounds, Tyler C. Thacker conceived the research, contributed to the design of the lab-scale testing platform from the bacterial safety aspects, oversaw bacterial culture growth, and preformed sampling, Somchai Rice assisted with the GC-MS and sample analysis, Jacek A. Koziel conceived and supervised the research, contributed to the design of the lab-scale testing platform, Pauline Nol and Kurt C. VerCauteren provided funding for this research and contributed to review of the manuscript. All authors have reviewed and given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maurer, D.L., Ellis, C.K., Thacker, T.C. et al. Screening of Microbial Volatile Organic Compounds for Detection of Disease in Cattle: Development of Lab-scale Method. Sci Rep 9, 12103 (2019). https://doi.org/10.1038/s41598-019-47907-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47907-w
This article is cited by
-
Micro-Chamber/Thermal Extractor (µ-CTE) as a new sampling system for VOCs emitted by feces
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.