Abstract
Total retocolectomy with ileal pouch-anal anastomosis (IPAA) is the surgery of choice for patients with ulcerative colitis (UC) that are refractory to clinical treatment. Pouchitis is one of the most common complications after this procedure. Defects in autophagy have been reported in inflammatory bowel diseases. However, there are no studies on the IP. Therefore, we studied markers for autophagy in the IP mucosa of UC and FAP patients comparing them to controls with a normal distal ileum. Sixteen patients with IP in “J” shape, asymptomatic and with endoscopically normal IP were evaluated. The control group consisted of eight patients with normal colonoscopy. There was a significant decrease in the transcriptional levels of ATG5, MAP1LC3A and BAX in the FAP group. There was also a decrease in the protein level of Beclin-1 in the UC and FAP compared to the control group. Although the LC3II levels by immunoblot were higher in the UC group, LC3/p62 co-localization were lower in the immunofluorescence analysis in the UC and FAP compared to the control group. Corroborating these results, there was an increase of p62 by immunoblot in the UC group. These findings indicated a modulation of macroautophagy markers in the IP, which may explain the mucosa inflammation predisposition.
Similar content being viewed by others
Introduction
Ulcerative colitis (UC) is a chronic intestinal inflammation that can affect the large intestine and rectum. Its etiology is not completely established. Familial adenomatous polyposis (FAP) is an autosomal dominant disease which affects young individuals and is associated with the formation of multiple polyps in the large intestine and rectum, which invariably implies a greater risk of cancer1,2. Both diseases, despite being different, may require the same surgical procedure. The ileal pouch-anal anastomosis (IPAA) is the elective procedure of choice in the surgical management of refractory UC, and FAP with many polyps in the rectum3. The main complication after this procedure is the pouch inflammation (pouchitis) that can affect up to 45 percent of patients who are submitted to IPAA for UC, and only five percent of the FAP patients who undergo the same procedure4. This suggests that constitutive differences between UC and FAP pouches have a critical role in its pathogenesis.
Pouchitis develops only after ileostomy closure, when the pouch mucosa starts to be exposed to the fecal stream5. The distinct immunological aspects of the different inflammatory bowel diseases (IBD), specifically UC, which involve impaired innate and adaptive responses, associated to genetic susceptibility, environmental factors, and intestinal microbiota may be involved in the pouch inflammation etiology5,6.
Autophagy is an evolutionarily conserved catabolic pathway that consists of selective degradation of cellular components and a homeostatic mechanism that protects cells exposed to stress situations (toxins, starvation)7,8. There are three primary forms of authophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA)9. Although there are no indications of genetic mutations related to the mechanism of autophagy associated to UC susceptibility, the transcriptional and protein evaluation of this mechanism in the ileal pouch mucosa is of fundamental relevance. Alterations of apoptosis in this tissue have already been described previously10 and both signaling pathways, autophagy and apoptosis, are interconnected11,12. Indeed, differential expression of Beclin1, a relevant protein that is involved in the initiation of autophagy, was already seen in the colon of UC patients4. Moreover, epigenetic alterations, which have recently been related to the etiology of IBD13,14, can determine transcriptional changes, which in turn help to better understand the mechanisms that predispose patients to the inflammatory process in the ileal pouch justifying its study. Thus, we evaluated molecules involved in the autophagy pathways in ileal pouch mucosa of UC and FAP patients, even in the absence of clinical, endoscopic and histological inflammation, in order to understand if there is underlying modulation in these pathways that could mediate molecular inflammation in the IP.
Results
Histological analysis to evaluate the Pouchitis Disease Activity Index
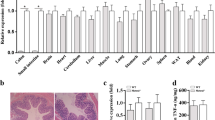
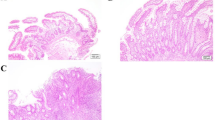
Among the aspects analyzed by the PDAI, the histological analysis of the biopsies collected in the ileal pouch mucosa of UC and FAP patients demonstrated a small amount of acute inflammatory cells infiltration, absence of crypts destruction, cell architecture preservation and presence of goblet cells, as demonstrated in Fig. 1B,C. Similarly, no significant histological changes were found in the biopsies performed in normal ileum (Control group) (Fig. 1A).
Haematoxylin and Eosin (H&E) staining of ileal pouch mucosa biopsy of representatives Familial Adenomatous Polyposis (FAP) and Ulcerative Colitis (UC) patients. (A) Ileal pouch mucosa of a normal control (CTR Group). (B) Ileal pouch mucosa of FAP patient (FAP Group). (C) Ileal pouch mucosa of UC patient (UC Group). (D) Polymorphonuclear (PMN) leukocyte number of the lamina propria in the CTR, FAP and UC Groups. There is no statistical difference among the groups. Nuclear counterstaining: Mayer’s haematoxylin. Original magnification X20.
The polymorphonuclear leukocyte count of the lamina propria is shown in the Fig. 1D. Photomicrographs were taken using a Leica DM 4500B microscope and Leica DFC 290 digital camera system with Leica Application Suite version 3.8 Software (Leica Microsystems, Wetzlar). Three fields for each sample were captured. The immune cells of the lamina propria were counted for quantitative analysis, which was analyzed by two blinded observers (N.M.P. and L.B.P.) in a panchromatic objective field of higher magnification 40X.
PDAI was performed for all patients taking into account clinical, endoscopic and histological aspects (see supplementary figure). All patients evaluated had PDAI < 7. The aim of our study was to analyze patients without inflammation to show if there were underlying molecular alterations in the ileal pouch mucosa, even in the absence of endoscopic and histological inflammation.
Transcriptional analysis of the autophagy related genes in the ileal pouches from fap and uc patients
Patients with FAP showed decreased mRNA levels of ATG5 in the ileal pouch when compared to UC (p < 0.01; Fig. 2D), and decreased levels of MAP1LC3A compared to the controls (p < 0.05; Fig. 2E). No differences were observed in the other genes (p > 0.05; Fig. 2A,B and C). To explain these findings, we decided to evaluate apoptosis related genes and although there was no statistical differences in BCL2 expression, an anti-apoptotic gene (p > 0.05; Fig. 2G), we found decreased BAX levels in the FAP group when compared to CTR group (p < 0.01; Fig. 2F). BAX encodes a pro-apoptotic protein, that when it is decreased, it leads to the inhibition of autophagy related genes, such as ATG5.
Evaluation of autophagy and apoptosis related gene expressions in the ileal pouch mucosa of Familial Adenomatous Polyposis (FAP) and Ulcerative Colitis (UC) patients. Transcriptional analysis reveals autophagy markers modulation in the ileal pouch mucosa of FAP patients. mRNA levels (qRT-PCR) of ULK1 (A), BECN1 (B), ATG16L1 (C), ATG5 (D), MAP1LC3A (E), BAX (F) and BCL2 (G) in ileal pouch mucosa of controls (CTR Group), FAP patients (FAP Group) and UC patients (UC Group). For FAP, n = 8; for UC, n = 8; for CTR, n = 8; *p < 0.05, **p < 0.01 and ***p < 0.001.
Transcriptional analysis of the autophagy related genes in the afferent limb of ileal pouches from fap and uc patients
There were no differences in the mRNA levels of autophagy related genes comparing the control, FAP-AF and UC-AF groups (p > 0.05; Fig. 3), which shows no transcriptional alterations among the afferent limbs and the normal terminal ileum mucosa.
Evaluation of autophagy gene expressions in the ileal pouch afferent limb mucosa of Familial Adenomatous Polyposis (FAP) and Ulcerative Colitis (UC) patients. Transcriptional analysis reveals no differences compared to the controls. mRNA levels (qRT-PCR) of ULK1 (A), BECN1 (B), ATG16L1 (C), ATG5 (D), and MAP1LC3A (E) in ileal pouch afferent limb mucosa of controls (CTR Group), FAP patients (FAP-AF Group) and UC patients (UC-AF Group). For FAP-AF, n = 8; for UC-AF, n = 8; for CTR, n = 8; *p < 0.05, **p < 0.01 and ***p < 0.001.
Protein analysis by immunoblotting revealed modulation of autophagy markers in the ileal pouch of uc and fap patients
In order to better evaluate the autophagy pathway, we measured the proteins amount by immunoblotting using the same samples of PCR analysis. We found decreased levels of Beclin-1 in FAP, UC-AF and UC groups when compared to control group (p < 0.05; Fig. 4A). Although LC3 level was increased in UC patients when compared to CTR and FAP-AF groups (p < 0.05; Fig. 4B), an increased non-degraded p62 was observed in the UC group (p < 0.05; Fig. 4C). Beclin-1 participates in the early stages of the autophagy pathway. However, p62 is a relevant molecule, which binds to LC3 and is responsible for carrying ubiquitinated unfolded proteins into the autophagosome and enable their degradation in the lysosome. p62 is an adapter protein, thus, if it is increased means failure in the degradation process, i.e. macroautophagy failure.
Ileal pouch mucosa of Familial Adenomatous Polyposis (FAP) and Ulcerative Colitis (UC) patients shows autophagy protein markers modulation. Western blot analysis of Beclin-1 (A), LC3 (B), p62 (C) and HSC-70 (D) in ileal pouch (FAP and UC Groups) and in its afferent limb mucosa (FAP-AF and UC-AF Groups) of FAP and UC patients compared to controls (CTR Group). Each band represents one patient. For FAP, n = 8; for UC, n = 8; for CTR, n = 8; for FAP-AF, n = 8; for UC-AF, n = 8; *p < 0.05, **p < 0.01 and ***p < 0.001. ASU: arbitrary scanning unit.
HSC-70 levels, which is a marker of chaperone-mediated autophagy, were similar among the groups (p > 0.05; Fig. 4D).
Immunofluorescence protein analysis confirmed modulation of autophagy markers in the ileal pouch of uc and fap patients
To confirm the protein-related autophagy expression data, co-localization for LC3 and p62 was performed.
Immunofluorescence staining of LC3 and p62 co-localization in the ileal pouch mucosa of Familial Adenomatous Polyposis (FAP) and Ulcerative Colitis (UC) patients. (A) Quantitative analysis of immunofluorescence staining for LC3 and p62 co-localized in FAP, UC and control (CTR) groups. (B) Representative staining of fixed paraffin-embedded ileal mucosa from the CTR, FAP and UC groups, showing low number of positive cells in FAP and UC groups compared to the CTR group. Positive cells are shown in orange (co-labeled by PI and FITC; overlay image) or red and green in the same cytosol (co-labeled by Alexa Fluor® 488 and Cy3®). Nucleus was stained with DAPI (blue-fluorescent). The arrows show the positive cells. Images were obtained using a 40X objective. For FAP, n = 8; for UC, n = 8; for CTR, n = 8; *p < 0.05, **p < 0.01 and ***p < 0.001.
Despite the increased LC3 II levels verified in the ileal pouch mucosa of UC patients by immunoblotting, the immunofluorescence analysis revealed a significantly lower number of LC3 and p62 co-localized cells in the FAP and UC groups, when compared to the CTR group (p < 0.05; Fig. 5A, quantitative analysis). Representative images of LC3 and p62 co-staining in the ileal pouch mucosa of normal distal ileum, FAP and UC patients are shown in Fig. 5B, where the positive cells are well identified in the different groups (orange/yellow in the cell cytoplasm; nucleus is counter-stained in blue).
Discussion
Previous studies have shown increased pro-inflammatory cytokines, nuclear transcription factor STAT-1, and bacterial antigen receptors such as TLR4 in the ileal pouch mucosa of UC patients, when compared to FAP and controls of normal distal ileum, even in patients without pouchitis15,16,17. These findings showed the greater susceptibility of UC patients to the inflammatory process in the ileal pouch after IPAA. In addition to those findings, a decrease in apoptosis in the ileal pouch mucosa of UC patients was also reported in the literature10. It is known that there is a close relationship between the mechanism of apoptosis and autophagy7,9,11. Hao X et al. verified increased levels of beclin-1 in colonic mucosa of UC patients18. However, there were no studies evaluating cellular autophagy, a relevant mechanism for recycling dysfunctional cellular components present in the cytoplasm, in the ileal pouch of UC and FAP patients.
There are three primary forms of authophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA)7,8,9,19. In macroautophagy, targeted cytoplasmic components are isolated from the rest of the cell within a double-membraned vesicle (autophagosome)20,21. The autophagosome can fuse with lysosomes and the proteins are degraded and recycled. On the other hand, microautophagy is mediated by direct lysosomal engulfment of the cytoplasmic components, which is trapped in the lysosome by the membrane invagination22. The recognition of the protein substrate by specific proteins such as chaperones in the cytosol, binding directly to the lysosome, translocating across it, without additional vesicles formation, is the characteristic of CMA23. For this reason, autophagy prevents the accumulation of abnormal proteins, is also involved in the genomic stability, and participates of the removal of intracellular pathogens24. When there is an autophagy deficiency, it promotes cytoplasmic protein inclusions, which are composed of misfolded proteins. The accumulation of deformed organelles can lead to cell injury and diseases8,25.
In the present study, the purpose was to study patients without pouchitis to demonstrate if there was underlying modulation of autophagy markers in the ileal pouch mucosa, even in the absence of endoscopic and histological inflammation. For this, we applied the PDAI (Pouchitis Disease Activity Index)26, and all patients had PDAI less than seven points. We showed increased LC3 II levels in the ileal pouch mucosa of UC patients by immunoblotting analysis. However, the decreased levels of beclin-1 and non-degraded p62 observed in Fig. 4A,C respectively, and also the less number of LC3/p62 co-localized cells in the UC group compared to controls reinforce the finding of autophagy markers defective modulation. Beclin-1 initiates the autophagy process. LC3 enroll in the autophagossome formation and p62 binds to LC3 and is responsible for carrying abnormal proteins into the autophagosome27. When p62 is accumulated in the cytoplasm, not binding to LC3, it signalizes that the macroautophagy may be deficient, even higher LC3 II levels is detected. Additionally, we did not verify altered CMA markers between the groups.
An interesting finding was also the detection of decreased autophagy markers in the ileal pouch mucosa of FAP patients when compared to controls. The transcriptional analysis showed fewer levels of ATG5 and MAP1LC3A in FAP group, besides decreased Beclin-1 protein levels by immunoblotting analysis, and finally, decreased number of total LC3 and LC3/p62 co-localized cells verified by immunofluorescence compared to control group. To explain these findings, we decided to evaluate apoptosis related genes. Although there were no statistical differences in BCL2 expression, we found decreased BAX level, which encodes a pro-apoptotic protein, in the FAP group when compared to controls. Decreased apoptosis markers were already described in the ileal pouch mucosa of FAP patients, what may explain the tendency to low cell turn over and possible development of adenomas in this syndrome10. The decreased BAX levels in FAP group can lead to inhibition of autophagy related genes, as ATG5, and may explain the decreased levels of proteins related to autophagy, as we showed in Figs 4 and 5.
Conversely, the defect autophagy in UC pouches may be explained by other mechanism. Increased levels of TLR4 were already observed in UC ileal pouch mucosa, even in the absence of endoscopic inflammation17. The relationship between TLR4 and immune system cells, mainly macrophages, frequently are associated to increased autophagy in those cells, especially after LPS treatment, which is a TLR4 agonist28,29. However, there is data from animal experimentation that addresses the role of autophagy in macrophage polarization30. Despite being in the context of obesity, it shows that in the obese animal, which exhibits increased TLR expression, autophagy is decreased in macrophages isolated from the peritoneum. They correlate this impaired autophagy with changes in the m1 and m2 macrophages profile, leading to inflammation. Therefore, in chronic stimuli such as obesity, autophagy is modulated in some tissues (hypothalamus, liver, muscle and macrophage)31,32,33,34,35. There are still no studies in intestinal mucosa addressing the negative regulation of autophagy through TLR activation, which may explain our results in UC ileal pouch mucosa. The mechanisms by which this may happen are still grounds for investigation in other tissues36. In fact, we verified decreased of macroautophagy markers in the ileal pouch mucosa of both, UC and FAP, but the mechanisms to explain may be distinct, analyzing data already published. In FAP, decreased autophagy markers may be related to impaired apoptosis, otherwise in UC, may be mainly due to increased TLR activation.
Therefore, autophagy is relevant to the cell survive, since the accumulation of unfolded and abnormal proteins leads to activation of pro-inflammatory pathways. Those evidences of autophagy markers modulation may explain the prone to inflammation in the ileal pouch mucosa, mainly in UC. However, some limitations should be considered. First, this is a descriptive cross-sectional observational study and did not intend to correlate with the occurrence of future clinical manifestations. For this purpose, we would need a longitudinal study with a larger cohort and long-term follow-up. Second, we did not measure the autophagic flux directly. However, the ubiquitin-associated protein p62, which binds to LC3, was used to monitor autophagic flux indirectly, as we did in Fig. 5. In addition, we applied several assays to confirm the results, and also to explain contradictory ones. These findings were the first to show modulation of autophagy markers in the ileal pouch of UC and FAP, even in patients without clinical and endoscopic inflammation.
This subject deserves further studies and detailed mechanisms37,38, which can help to find out new targets to ameliorate inflammation in the ileal pouch and even in UC. If these findings are confirmed in a longitudinal study, exploring the correlations with clinical settings, then this work provides novel insight into the complex pathogenesis of primary pouch inflammation.
Methods
Mucosal biopsies were obtained from eight patients with non-inflamed IPAA after rectocolectomy for UC [median age, 52 (range, 38–66) years; 75% male; 25% female], and eight patients with non-inflamed IPAA after rectocolectomy for FAP [median age, 52.5 (range, 35–70) years; 37.5% male; 62.5% female]. Biopsies of the intestinal mucosa of these patients were collected from the ileal pouch (UC and FAP Groups) and from the afferent limb of the ileal pouch (UC-AF and FAP-AF Groups). The postoperative follow-up was 186.5 (13–360) months. The reservoir design was of the “J” type, and the right colon vascular arcade was preserved as a supplementary blood supply to the terminal ileum39. All the patients in this study had the absence of pouchitis, which was defined clinically, histology and endoscopically, according to the PDAI (Pouchitis Disease Activity Index)26 and the ileostomy closed for more than one year. In the control group (CTR Group), eight individuals with normal colonoscopy examination were included, with a median age of 62.5 (range, 53–72) years and 37.5% were female. Six biopsies from each patient were obtained from the terminal ileum (control), from the afferent limb and from the ileal pouch (UC and FAP).
This study was approved by the Ethics Committee of the University of Campinas (UNICAMP), all patient signed the informed consent form, and was performed in accordance with the Declaration of Helsinki. The study was carried out at the University of Campinas, IBD Research Laboratory of the Surgery Department, and at the Laboratory of Cell Signaling of the Internal Medicine Department.
Histological analysis (hematoxylin - eosin)
Biopsies were embedded in paraffin blocks for histological analysis. Sections of 5 μm were cut and stained with hematoxylin and eosin dye. Photomicrographs were taken using a Zeiss Axiophot microscope and Cannon Power Shot G5 digital camera system (Cannon Inc., Tokyo). Fields of higher magnification (20X) were scanned and random fields were analysed. The histological part of the PDAI was performed.
RT-PCR Analysis
Biopsies from the mucosa of the terminal ileum and from the UC and FAP patients (afferent limb and ileal pouch) were snap-frozen in liquid nitrogen and stored at −80 °C until use. Total RNA was extracted using Trizol (Invitrogen), according to the manufacturer’s instructions. RNA purity and concentration were determined by UV spectrophotometry at 260 nm. RNA was reverse transcribed using oligo (dT) primers and reverse transcriptase (High-Capacity cDNA Reverse Transcription™ Kit, Applied Biosystems). The reaction mixture (20 µl) was incubated at 42 °C for 60 min, then for 10 min at 70 °C, and cooled on ice. RT-PCR was performed on resulting cDNA, using the manufacturer’s protocol, in a 25 µl reaction volume per capillary. Gene-specific primers (TaqManTM - Applied Biosystems™) were ATG16L1 (Hs00250530_m1), MAP1LC3A (Hs00261291_m1), BECN-1 (Hs00186838_m1), ATG5 (Hs00169468_m1), ULK1 (Hs00177504_m1), BCL2 (Hs00608023_m1), BAX (Hs00180269_m1) and GAPDH (NM_002046.3). RT-PCR amplification consisted of an initial denaturation step (50 °C for 2 min and 95 °C for 10 min), 40 cycles of denaturation (95 °C for 15 s), annealing (53 °C for 20 s) and extension (72 °C for 20 s), followed by a final incubation at 60 °C for 1 min. All measurements were normalized by the expression of GAPDH gene, considered as a stable housekeeping gene40.
Real-time PCR analysis of gene expression was performed in a STEP ONETM Real-Time PCR System (Applied Biosystems). The optimal concentration of cDNA and primers, as well as the maximum efficiency of amplification, were obtained by five-point, two-fold dilution curve analysis for each gene. Real-time data were analyzed using the Sequence Detector System 1.7 (Applied Biosystems). Reagents for Real-time PCR analysis were from Invitrogen (Carlsbad, CA, USA) and Applied Biosystems (Foster City, CA, USA).
Immunoblotting – Gel electrophoresis
Biopsies were snap-frozen in liquid nitrogen and stored at −80 °C until use. For total protein extract preparation, the fragments were homogenized in solubilizing buffer at 4 °C [1% Triton X-100, 100 mM Tris-HCl (pH 7.4), 100 mM sodium pyrophosphate, 100 mM sodium fluoride, 10 mM EDTA, 10 mM sodium orthovanadate, 2.0 mM phenylmethylsulfonyl fluoride (PMSF), and 0.1 mg aprotinin/ml] with a Polytron PTA 20 S generator (model PT 10/35; Brinkmann Instruments, Westbury, NY) operated at maximum speed for 30 sec. Insoluble material was removed by centrifugation (12000 rpm at 4 °C for 40 min). The protein concentration of the supernatants was determined by BCA method (PierceTM BCA Protein Assay Kit. Catalog number 23225). Aliquots of the supernatants containing 50 μg total proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes and blotted with indicated antibodies as described in the results. Specific bands were labeled by a chemiluminescence reaction (SuperSignal West Pico Chemiluminescent Substrate from Pierce Biothecnology, Inc. Rockford, IL) and quantified by optical densitometry (Un-Scan-It Program). We have applied Ponceau staining to check equal loading of gels and membrane transfer (see supplementary information)41,42.
All the reagents for SDS-polyacrylamide gel electrophoresis and immunoblotting were from Bio-Rad Laboratories (Richmond, CA, USA). HEPES, phenylmethylsulfonyl fluoride, aprotinin, dithiothreitol, Triton X-100, Tween 20, glycerol, and BSA (fraction V) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Nitrocellulose paper (BA85, 0.2μm) and the reagents for chemoluminescence protein labeling in immunoblots were purchased from Amersham (Aylesbury, UK). The primary antibodies against Beclin (ab-16998), p62 (ab56416 or ab91526) were from AbCam, Cambridge, MA, USA; and LC3 (#2775) from Cell Signaling, Boston, MA, USA or LC3 (ab48394) from AbCam, Cambridge, MA, USA, and Hsc70 (sc7298) from Santa Cruz Biotechnology, Santa Cruz, CA, USA. The protein molecular weight was assessed by the PageRulerTM from Fermentas (Glenburnie, MD).
LC3 and p62 Immunofluorescence staining
Histological sections of 5μm were also performed for immunofluorescence procedures of samples included in paraffin blocks. The preparation of slides was performed (deparaffinization and hydration), followed by antigen retrieval. The tissue was incubated in primary antibody anti-LC3 (M115–3) from MBL Internacional Corporation Woburn, MA, USA with a dilution of 1:500 at 4 °C overnight and after with secondary antibody conjugated with Goat Anti-Mouse IgG H&L (Cy3®) preadsorbed (ab97035) from AbCam, Cambridge, MA, USA, with a dilution of 1:500 at room temperature for 1 hour. Three fields for each sample were captured and analyzed through the Leica confocal LAS AF Lite Version 2.6 Software (Leica Microsystems, Wetzlar). All cell type stained in the cytosol for Cy3® were considered positive for quantitative analysis, which was performed by ImageJ2, by percentage of LC3 per total tissue area in a panchromatic objective field of higher magnification 40X43.
Indeed, we evaluated the autophagy by assessing LC3 and p62 co-localization44. The tissue was incubated in primary antibody anti-LC3 (M115–3 from MBL Internacional Corporation Woburn, MA, USA) and anti-p62 (ab91526 from AbCam, Cambridge, MA, USA) with a dilution of 1:500 at 4 °C overnight. The secondary antibody was Alexa Fluor® 488 (goat anti-rabbit IgG H&L: ab150077 from AbCam, Cambridge, MA, USA) in a dilution of 1:1000 at room temperature for 1 hour or Goat Anti-Mouse IgG H&L (Cy3®) preadsorbed (ab97035 from AbCam, Cambridge, MA, USA), with a dilution of 1:500 at room temperature for 1 hour. DAPI was used for nuclear staining. Three fields for each sample were captured and analyzed through the Leica confocal LAS AF Lite Version 2.6 Software (Leica Microsystems, Wetzlar). All cell type stained in the cytosol for Alexa Fluor® 488 and Cy3® were considered positive for quantitative analysis, which was performed by ImageJ2, by the percentage of co-localized pixel volume in a panchromatic objective field of higher magnification 40X43.
Statistical analysis
All results were reported as means ± SEM. Data were analyzed by non-parametric Test, comparing all groups. The level of significance was set at p < 0.05.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the University of Campinas (UNICAMP), all patient signed the informed consent form, and was performed in accordance with the Declaration of Helsinki.
Consent for publication
Ethical approval by the ethic board of the University of Campinas (UNICAMP) and consent of patients are included in the original publications.
Availability of data and materials
All data generated or analyzed during this study are included in this article and in the additional files.
References
Scaldaferri, F. & Fiocchi, C. Inflammatory bowel disease: Progress and current concepts of etiopathogenesis. J. Dig. Dis. 8(4), 171–178 (2007).
Half, E., Bercovich, D. & Rozen, P. Familial adenomatous polyposis. Orph. J. Rare Dis. 4(1), 22 (2009).
McGuire, B., Brannigan, A. & O’Connell, P. Ileal pouch–anal anastomosis. Br. J. Surg. 94(7), 812–823 (2007).
Lovegrove, R. et al. A Comparison of Adverse Events and Functional Outcomes After Restorative Proctocolectomy for Familial Adenomatous Polyposis and Ulcerative Colitis. Dis. Colon Rectum 49(9), 1293–1306 (2006).
Shen, B. Acute and chronic pouchitis-pathogenesis, diagnosis and treatment. Nature Rev. Gastroenterol. Hepatol. 9(6), 323–333 (2012).
Hata, K., Watanabe, T., Shinozaki, M. & Nagawa, H. Patients with extraintestinal manifestations have a higher risk of developing pouchitis in ulcerative colitis: multivariate analysis. Scand. J. Gastroenterol. 38(10), 1055–1058 (2003).
Kang, R., Zeh, H. J., Lotze, M. T. & Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18(4), 571–80 (2011).
Yang, Y. P., Liang, Z. Q., Gu, Z. L. & Qin, Z. H. Molecular mechanism and regulation of autophagy. Acta Pharmacol. Sin. 26(12), 1421–34 (2005).
Codogno, P. & Meijer, A. J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12(2), 1509–18 (2005).
Leal, R. F. et al. Detection of epithelial apoptosis in pelvic ileal pouches for ulcerative colitis and familial adenomatous polyposis. J. Trans. Med. 8(1), 11 (2010).
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8(10), 1124–1132 (2006).
Zhou, F., Yang, Y. & Xing, D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 278(3), 403–413 (2010).
Wang, M. et al. Gene–gene and gene–environment interactions in ulcerative colitis. Human Gen. 133(5), 547–558 (2014).
Wang, M. et al. A Novel Approach to Detect Cumulative Genetic Effects and Genetic Interactions in Crohn’s Disease. Inflamm. Bowel Dis. 19(9), 1799–808 (2013).
Leal, R. F. et al. Differential expression of pro-inflammatory cytokines and a pro-apoptotic protein in pelvic ileal pouches for ulcerative colitis and familial adenomatous polyposis. Tech. Coloproctol. 12(1), 33–8 (2008).
Leal, R. F. et al. Activation of signal transducer and activator of transcription-1 (STAT-1) and differential expression of interferon-gamma and anti-inflammatory proteins in pelvic ileal pouches for ulcerative colitis and familial adenomatous polyposis. Clin. Exp. Immunol. 160(3), 380–5 (2010).
Paiva, N. M. et al. Differential expression of TLR2, TLR4 and JNK in mucosa of ileal pouches for ulcerative colitis. Is there a role for bacterial antigen pathway in asymptomatic patients? Int. J. Clin. Exp. Med. 4, 179–186 (2011).
Hao, X., Yang, B., Liu, X., Yang, H. & Liu, X. Expression of Beclin1 in the colonic mucosa tissues of patients with ulcerative colitis. Int. J. Clin. Exp. Med. 15, 21098–105 (2015).
Tanida, I. Autophagy basics. Microbiol. Immunol. 55(1), 1–11 (2011).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Ann. Rev. Cell Dev. Biol. 27(1), 107–132 (2011).
Xie, Z. & Klionsky, D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9(10), 1102–9 (2007).
Li, W. W., Li, J. & Bao, J. K. Microautophagy: lesser-known self-eating. CMLS. 69(7), 1125–36 (2012).
Kaushik, S. & Cuervo, A. M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 22(8), 407–17 (2012).
Van Limberg, J., Stevens, C., Nimmo, E. R., Wilson, D. C. & Satsangi, J. Autophagy: from basic science to clinical application. Mucosal Immunol. 2(4), 315–330 (2009).
Komatsu, M. & Ichimura, Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 584(7), 1374–8 (2010).
Sandborn, W. J., Tremaine, W. J., Batts, K. P., Pemberton, J. H. & Phillips, S. F. Pouchitis after ileal pouch-anal anastomosis: a Pouchitis disease activity index. Mayo Clin. Proc. 69, 409–415 (1994).
Bjorkoy, G. et al. Monitoring autophagic degradation of p62/SGSTM1. Methods Enzymol. 452, 181–197 (2009).
Xu, Y. et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 27(1), 135–44 (2007).
Into, T., Inomata, M., Takayama, E. & Takigawa, T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 24(6), 1150–62 (2012).
Liu, K. et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 11(2), 271–84 (2015).
Portovedo, M. et al. Saturated fatty acids modulate autophagy’s proteins in the hypothalamus. PLoS One. 18 10(3), e0119850 (2015).
Meng, Q. & Cai, D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway. J. Biol. Chem. 16, 32324–32 (2011).
Yang, L., Li, P., Fu, S., Calay, E. S. & Hotamisligil, G. S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 9 ;11(6), 467–78 (2010).
Yan, H., Gao, Y. & Zhang, Y. Inhibition of JNK suppresses autophagy and attenuates insulin resistance in a rat model of nonalcoholic fatty liver disease. Mol. Med. Rep. 15(1), 180–186 (2017).
Potes, Y. et al. Overweight in elderly people induces impaired autophagy in skeletal muscle. Free Radic. Biol. Med. 23, 31–41 (2017).
Hu, N. & Zhang, Y. TLR4 knockout attenuated high fat diet-induced cardiac dysfunction via NF-κB/JNK-dependent activation of autophagy. Biochem. Biophys. Acta. 4439(17), 30015–7 (2017).
Macias-Ceja, D. C. et al. Autophagy stimulation prevents intestinal mucosal inflammation and ameliorates murine colitis. Br. J. Pharmacol. 174(15), 2501–2511 (2017).
Huett, A. & Xavier, R. J. Autophagy at the gut interface: mucosal responses to stress and the consequences for inflammatory bowel diseases. Inflamm. Bowel Dis. 16(1), 152–74 (2010).
Góes, J. R. N., Coy, C. S. R., Amaral, C. A., Fagundes, J. J. & Medeiros, R. R. Superior mesenteric artery syndrome as a complication of ileal pouch-anal anastomosis. Dis. Colon Rectum. 38, 543–544 (1995).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25(4), 402–8 (2001).
Romero-Calvo, I. et al. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal Biochem. 401(2), 318–20 (2010).
Fortesa, M. A. S. et al. Housekeeping proteins: How useful are they in skeletal muscle diabetes studies and muscle hypertrophy models? Analytical Biochemistry 504(1), 38–40 (2016).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 7–8 (2015).
Pugsley, H., Friend, S., Kong, R., Vaidyanathan, S. & Morrissey, P. Measuring autophagic flux by assessing LC3, p62 and LAMP1 co-localization using imaging flow cytometry. J. Immunol. 194, 206–18 (2015).
Acknowledgements
We are grateful to Prof. Tristan Torriani for English grammar revision. We thank Francesca Ramos and José Diego Botezelli for technical assistance. We also thank to the staff of the Life Sciences Core Facility (LaCTAD) from State University of Campinas (UNICAMP), for the Cell Biology analysis.This work was supported by FAPESP (São Paulo Research Foundation). Leandro Minatel Vidal Negreiros (co-author) received scholarship from FAPESP.
Author information
Authors and Affiliations
Contributions
M.M. and R.F.L. designed the study; N.M.P., M.L.S.A. recruited patients; L.M.V.N., M.L.S.A. and C.S.R.C. assessed clinical disease activity; N.M.P., L.B.P., M.P. and A.C., performed the experiments; L.B.P. and L.M.V.N. carried out statistical analysis; and R.F.L., N.M.P., and M.M. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paiva, N.M., Pascoal, L.B., Negreiros, L.M.V. et al. Ileal pouch of ulcerative colitis and familial adenomatous polyposis patients exhibit modulation of autophagy markers. Sci Rep 8, 2619 (2018). https://doi.org/10.1038/s41598-018-20938-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20938-5
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.