Abstract
Barley genomic resources are increasing rapidly, with the publication of a barley pangenome as one of the latest developments. Two-row spring barley cultivars are intensely studied as they are the source of high-quality grain for malting and distilling. Here we provide data from a European two-row spring barley population containing 209 different genotypes registered for the UK market between 1830 to 2014. The dataset encompasses RNA-sequencing data from six different tissues across a range of barley developmental stages, phenotypic datasets from two consecutive years of field-grown trials in the United Kingdom, Germany and the USA; and whole genome shotgun sequencing from all cultivars, which was used to complement the RNA-sequencing data for variant calling. The outcomes are a filtered SNP marker file, a phenotypic database and a large gene expression dataset providing a comprehensive resource which allows for downstream analyses like genome wide association studies or expression associations.
Similar content being viewed by others
Background & Summary
Barley is one of the most important crops worldwide (5th in 2020 on area harvested, FAOSTAT1) and has a high value in the European agricultural sector underpinning the beer and whisky industries2. New barley cultivars are introduced to the market every year, after being evaluated for multiple traits e.g., disease resistance, yield, and malting quality traits3. Barley breeding and the introduction of barley cultivars started at the beginning of the 19th century in the UK and by the end of the 19th century all over Europe4. Instead of seeds being grown by the farmer with some saved for subsequent sowing the following year, breeding institutes were established, with the mission to develop improved seed stocks. Early cultivars were developed through mass selection and later followed by line selection from landraces. Initial breeding efforts focused on increasing yield5. Due to the considerable success of these breeding efforts, seed stocks soon became distributed across the continent and each country started their own breeding program by incorporating local landraces in crosses with these generally higher yielding genotypes. This cross-breeding technique of simple crosses followed by selection quickly led to an increase in yield as shown for spring barley in Germany with a doubling of yield from 1800 to 19006. Breeding developed further by intentionally mutating seeds with chemicals or radiation to induce higher genetic variation in the offspring7. One of the most notable results from mutation breeding were the dwarfing genes which were critical for the green revolution8. Shorter stature cultivars provided the advantage of preventing lodging which was crucial for the development of high-yielding cultivars with heavy spikes. Complementing traditional to cross- and mutation-breeding, molecular technologies developed further and were quickly adopted. One of the most successful advances was marker-assisted selection (MAS) which deploys molecular markers to detect allelic variations within a genome. The most common markers used in breeding nowadays are single nucleotide polymorphisms (SNPs)9. MAS is used for rapid and high-throughput selection of new genotypes and has matured from single marker analysis to genome-wide selection approaches. While SNPs are a key component of the genotyping platforms used in plant breeding purposes, they can also be used for gene discovery. Quantitative trait locus (QTL) mapping and genome wide association studies (GWAS) are valuable to identify alleles for genes underpinning genetically complex traits10,11,12,13. High throughput genetic markers are however only one of a number of genetic and genomic resources that have effectively revolutionised genetics and breeding. Next generation sequence data formed the basis of the first linear barley genome published in 2017 from the cultivar Morex14 which has been followed quickly by additional genomes from other cultivars15. The availability of “reference genome sequences” has both simplified the process and allowed a more precise identification of the causative genes controlling phenotypic traits.
Here we introduce new genetic and genomic datasets assembled from a European two-row spring barley population that is representative of pan-European breeding progress across the years from 1830 to 2014. A total of 209 50 K SNP-array16 genotyped barley cultivars were selected and grown in replicated field trials across three contrasting environments and for two years to score agronomic traits. Six different tissues from each cultivar were harvested and RNA was isolated for the collection of tissue and genotype specific transcript abundance (RNA-seq) data. Using both this RNA-seq data and whole genome shotgun sequence data from all individuals in the population, an exhaustive collection of high confidence SNP markers was assembled. We describe these datasets and provide examples of how they can be used.
Methods
Barley material and field trials
We assembled a collection of 209 European two-row spring barley cultivars (Supplemental Table 1), which is a representative subset of previously described two-row spring European barley populations10,11,17,18,19 that show a significant increase in yield over time. A small number of seed are available on request from the corresponding author and after signing a Standard Material Transfer Agreement (SMTA). Pedigree data was collected from publications17,20, and the following two websites: https://grinczech.vurv.cz/gringlobal/search.aspx and https://www.lfl.bayern.de/mam/cms07/ipz/dateien/abst_gerste.pdf. Field experiments were conducted at the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben, Germany, the James Hutton Institute (JHI) in Dundee, UK and the University of Minnesota (UMN) in St. Paul, USA in 2019 and 2020. At IPK and UMN, 100 grains of each genotype were sown in 1 m long double-rows in a completely random design with three replications in both years. At JHI, a seed density estimated to produce 350 plants per m2 for plot sizes of 2 m × 1.5 m was established. In 2019 a single replicate was grown and in 2020 a completely random design with two replicates. In addition, a polytunnel trial was included at JHI in 2019. Plant material was grown in 7 litre sized pots, 4 seeds per pot, in 3 replicate sets in a completely random design. Each replicate set had 8 columns and 30 rows and contained a replicate of each of the 209 genotypes.
Phenotyping
In total, 29 phenotypes were recorded on a per-plot basis in the field trials or on a per-pot basis in the polytunnel experiment. Developmental traits, growth habit and plant height measurements were recorded in the trials as described in Table 1. To measure spike and grain traits, ten to 15 main tiller spikes were harvested at full maturity (Zadoks stage 92) per plot, excluding the outermost plants of each row to avoid edge effects. After recording of all spike traits, spikes were hand-threshed, and grains were subjected to size and weight measurements on a Marvin SeedAnalyzer 6 (MARViTECH GmbH, Germany). Samples were first weighted and then added on to the Marvin tray for optical measurements of the grain size.
Tissue sampling for RNA-seq
Six different tissues were sampled for RNA-seq analysis: crown, root, inflorescence, peduncle, spikelet and grain. For each tissue one RNA-seq sample per genotype was generated. At UMN, crown and root tissues were sampled from seven-day-old seedlings (GRO:0007060, first leaf unfolded). Ten seeds per genotype were surface sterilized and planted in moist vermiculite in individual Cone-tainers (6000 RLC3 size, Ray Leach, Tangent, OR). The Cone-tainers were put into a dark cold room for four days to achieve more consistent germination. Then they were moved into a growth chamber at 20 °C with 16 hours of light for seven days. Tissues were harvested within three hours, starting at 9:00 am USA Central Time Zone to reduce the circadian effect on gene expression. Roots were sampled by cutting the longest root from each seedling adjacent to the germinated seed, and crowns by removing the roots and keeping the 1 cm shoot tissue immediately above. For each individual genotype five plants were combined and snap frozen in liquid nitrogen.
At JHI, the barley plants grown in 2019 under polytunnel conditions were used for tissue sampling. When plants reached the booting stage, which was 84–85 days after germination, 3 – 5 cm whole developing inflorescence tissue was taken, two from each replicate per genotype per sample. Whole peduncles were taken at 2–5 cm in length, three from each replicate per genotype per sample when plants were 88–90 days old. Sampling took place in a two-hour period between 10:00 and 12:00 Western European Summer Time and samples were snap frozen in liquid nitrogen and stored at −80 °C.
At IPK, barley plants grown in the 2019 field trial were monitored daily by dissecting single spikelets and recording the date of green anther stage and flowering stage on paper tags attached to the spikes. Sampling was limited to a two-hour period between 10:00 and 12:00 am Central European Summer Time each day to reduce the circadian effect on gene expression. Three spikes per plot of one repetition were selected at green anther stage and two central spikelets from the centre of each spike were sampled. At 5 days post anthesis, three spikes per plot of one repetition were selected and six developing grains per spike were sampled from the central region of each spike. All samples were snap-frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
RNA extraction and RNA sequencing
RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with all buffers provided and treated with DnaseI following the manufacturer’s instructions. Buffer RLC was used for seedling root extractions, and Buffer RLT was used for all other tissue extractions. To ensure a high purity of spikelet and grain samples, a more rigorous cleanup using 700 µl RW1 and three wash steps with RPE was performed. The integrity of samples was determined using an Agilent 2100 Bioanalyzer, an Agilent 4200 TapeStation or a 1% agarose gel. All tested samples had a RNA integrity number (RIN) factor of > = 8 and were suitable for further processing. Paired-end libraries were constructed from spikelet and grain samples (IPK Gatersleben) and seedling root and crown samples (University of Minnesota Genomics Center, Minneapolis, MN, USA) using the Illumina TruSeq Stranded Total RNA Library Prep Plant with Ribo-Zero Plant kit and sequenced on the NovaSeq 6000 platform with a read length of 150 bp. For the inflorescence and peduncle samples (JHI) Illumina RNA-seq library preparation and RNA-seq was carried out by Novogene (Company Limited, Hong Kong). The libraries were prepared using NEBNext® Ultra™ Directional RNA Library Prep Kit and sequenced using Illumina NovaSeq 6000 (PE 150).
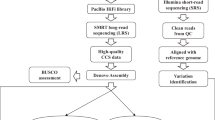
Bioinformatics
Read quantification
We generated 77.95 billion raw reads from RNA-seq of the six different tissues (Supplemental Table 2). Raw reads were trimmed with Trimmomatic 0.3921 to remove adapters and reads shorter than 60 bp. Salmon 1.3.022 was used for expression quantification including the gcBias setting to align trimmed reads to the transcriptome. We followed the approach of selective alignments by generating a decoy-aware transcriptome from the barley reference transcript dataset V2 (BaRTv2)23 and the reference genome of cv Barke15. This approach is recommended24 to reduce inaccurate transcript quantification caused by unannotated genomic loci that have a high sequence similarity to annotated transcripts.
Expression analysis
Tissue-specific genes were identified using different R packages25. For each tissue the raw counts were imported and combined to gene expression counts using tximport26. Raw counts were normalised (calcNormFactor), and log transformed to counts per million (cpm) using edgeR27. The tissue-specific expressed genes were identified by filtering for an average cpm of above 1 across all samples in this tissue and an average cpm of below −1 for all the other tissues. In addition, gene expression for two and more tissues were filtered with the same parameters to build the intersection sets required to create an UpSet28,29 plot of expressed genes in the different tissues. Gene ontology (GO) enrichment for the identified genes and visualisation were done as previously described30.
Variant calling
For variant calling, the trimmed RNA-seq reads were mapped to the reference genome of cv Barke15 using the two-pass mode implemented in STAR v. 2.7.531 allowing 6% mismatches normalized to read length, intron lengths between 60 and 15000 bp, a maximum distance of 2000 bp between mates and a maximum number of 30,000 transcripts per window. Due to the high number of reads in the grain and spikelet tissue the splice junction files were filtered for at least one uniquely-mapped read in more than one sample, with non-canonical splice sites removed and then used to generate a new genome index for the second mapping run. For the other tissues, the splice junction files from the first pass were provided as part of the input for the second mapping step. Duplicated reads were marked with Picard 2.18.2932 followed by filtering with bamtools 2.5.133 to remove reads with > = 2% mismatches and a mapping quality < = 50.The legacy algorithm of Freebayes 1.3.234 was used to call variants with a minimum fraction of alternate allele observations of 20%, a minimum alternate allele count of 2, a minimum coverage of 4, and minimum base and mapping qualities of 30.
Whole genome shotgun (WGS) approach
DNA was extracted from snap-frozen second leaves of greenhouse-grown (21 °C/18 °C day/night temperature) two-week old seedlings using a guanidinium thiocyanate-NaCl-based method as described35. DNA quality and quantity were assessed by agarose gel electrophoresis. The Nextera DNA kit (Illumina) was used for constructing libraries which were multiplexed and sequenced on a NovaSeq 6000 platform at IPK Gatersleben to generate 150-bp paired-end reads. A total of 12.16 billion raw paired-end reads (Supplemental Table 3) were trimmed with Cutadapt 1.1536 to remove adapters and reads shorter than 30 bp. Trimmed reads were mapped to the reference genome of cv Barke using Minimap2 2.1137. The resulting alignment files were sorted and duplicate-marked using Novosort 3.06.0538 and converted to cram files using samtools 1.839,40. On average the coverage was 4x across all samples with the lowest at 1.5x to the highest at 6.5x coverage (Supplemental Table 3). The ‘call’ function of Bcftools40 was used to call variants using genotype likelihoods calculated from alignments with a minimum quality score of 20 with the ‘mpileup function of BCFtools. Variants were re-called based on read depth ratios using a custom awk script similar to the one at https://bitbucket.org/ipk_dg_public/vcf_filtering/src/master/ with the following parameters modified: dphom = 1, dphet = 2, minhomn = 10, tol = 0.249, minmaf = 0.1, minpresent = 0.01.
Genotype marker file
The final genotype file was generated by filtering and merging multiple files. First all RNA-seq vcf files from the six tissues were filtered to remove insertions and deletions (Indels). SNPs corresponding to the robust BOPA markers50 were extracted from all six RNA-seq files. Pearson correlation between the markers and RNA seq files was calculated and VCFtools v0.1.16 with the parameter--diff-in-site was used to identify identical variants between the sets and those which differed. These two methods allowed for the identification of switched samples, those which did not correlate and SNPs which were inconsistent across the datasets. Swapped samples were renamed and those which did not correlate removed from further analyses. In addition, samples with a high number of heterogeneous SNPs (above 10%) were removed. RNA-seq SNPs from individual tissues were then merged, prioritizing homozygous calls while retaining heterozygous calls only if no homozygous calls were present in any of the tissues. After merging the six RNA-seq SNP datasets, the other two datasets first the WGS SNPs followed by the 50 K array SNPs16 were compared and added in the same manner. Calculating the Pearson correlation coefficient between the datasets and running VCFtools with the parameter --diff-in-site, removing or swapping samples if applicable.
The resulting unfiltered dataset contained 209 cultivars and 32,484,981 bi-allelic SNPs. The merged SNP dataset was filtered using TASSEL541 to remove SNPs with more than 20% missing data, minor allele frequency (MAF) < 0.01, heterozygosity >0.02, and only keeping bi-allelic SNPs. Missing data was imputed using the FILLIN plugin42 in TASSEL5 by first identifying haplotypes. For haplotype identification each chromosome was split into 500 blocks. The number of markers per haplotype block (-hapSize) was the total number of markers per chromosome divided by 500 and rounded to be divisible by 64 (TASSEL5 software requirement). Haplotypes were identified for each block with a maximum number of haplotypes of 20 (-maxHap 20) and at least five different genotypes per haplotype (-minTaxa 5). Haplotype information was used as input for the imputation. Further filtering removed seven lines that had more than 30% missing data after imputation (Aramir, Balder J, Dallas, KWS Irina, Power, Proctor and Spey), and one line was removed that had more than 2% heterozygosity (Rika). In a last filtering step, we removed SNPs which still had more than 20% missing data, MAF < 0.025 or heterozygosity > 0.02. SNPs were LD pruned with PLINK (v1.9)43 using a window size of 5000, a step size of 50 and an r2 threshold of 0.99. The final SNP dataset after pruning contained 201 cultivars and 1,509,447 SNPs. In the final SNP file 0.25% of markers represented markers from the 50k array, 25.1% from the RNA-seq data and 98.5% from the WGS data. The overlap between RNA-seq and WGS data is considerable with 98.6% of the RNA-seq markers also being identified by the WGS dataset.
Variant effect using SnpEff
To identify the effect of variants on the protein, we filtered the raw vcf files in a different way to generate an input file for SnpEff44. The aim for the genotype marker set explained above was to reduce the number of SNPs with pruning to a size which can be used for association analysis. For the variant effect we needed all the available SNP information and more importantly did not want to lose any SNPs due to pruning in gene space. For SNPs, the merged unfiltered vcf file containing RNA-seq, WGS and 50k data was filtered by removing heterozygous calls, removing SNPs with missing data in more than 20% of the samples and a minor allele frequency of <0.025. In addition, a dataset containing Indels was created by using the six vcf output files from the RNA-seq data after variant calling with Freebayes. All were filtered to keep Indels only, remove heterozygous calls, remove variants with missing data in more than 20% of the samples and a MAF of <0.025. The six Indel vcf files were combined into one. A SnpEff database was built based on BaRTv2 and the Barke reference genome.
Statistical analysis of phenotypic data, calculation of Best linear unbiased predictions (BLUPs) and heritability
Statistical analysis was performed using R 3.6.125. The Pearson correlation coefficient between experiments was calculated for each phenotypic trait and datasets showing an insignificant correlation (p > 0.05) with at least one other dataset of the same trait were removed before calculating BLUPs. The datasets being used in each of the BLUP calculations are listed in Supplemental Table 4. BLUPs were calculated across experiments using a randomized complete block model in META-R with experiments set as a random factor following formula 3 in Alvarado et al.45.
Genome wide association studies (GWAS)
Association between phenotype and genotype was done using the Mixed Linear Model (MLM)46 with GAPIT (version 3)47. As input, we used the genotype marker file of 201 cultivars and 1,509,447 SNPs. The BLUP values of Awn length were used to provide an example of the process. Three principal components (PCs) were calculated within GAPIT and model selection set to TRUE to enable GAPIT to select the optimal number of PCs for the individual phenotype based on a Bayesian information criterion (BIC).
Data Records
All raw data files for both raw RNA sequencing data and whole genome shotgun data have been deposited at the European Nucleotide Archive (ENA) under the following project number: PRJEB4906948 for the RNA-sequencing reads and PRJEB4890349 for the whole genome shotgun sequencing reads.
Phenotypic data and the SNP marker file are available through Germinate50: https://ics.hutton.ac.uk/germinate-barn/
The database contains the raw data by year and by site plus the calculated BLUP dataset.
The SNP marker file has been deposited in the European Variant Archive51 under the following project number: PRJEB6587552.
Derived datasets are available through e!Dal53 with the following https://doi.org/10.5447/ipk/2023/15 The datasets consist of two sets of gene expression files per tissue; one for the raw read counts and one for the TPM values mapped against BaRTv2. In addition, the data has been uploaded to ArrayExpress54 with the following accession numbers: E-MTAB-13231 (spikelet tissue)55, E-MTAB-13236 (grain tissue)56, E-MTAB-13235 (root tissue)57, E-MTAB-13234 (crown tissue)58, E-MTAB-13233 (inflorescence tissue)59 and E-MTAB-13232 (peduncle tissue)60.
Two further files containing variant identification are available through e!Dal. The first contains the SNPs and the second the Indel information with SnpEff annotation.
Technical Validation
Population
The 209 two-row spring barley population was selected from previously established datasets containing 647 cultivars10,11,17,18,19. To include a wide range of genetically representative individuals, we used available BOPA SNP data (as previously described)61 and did a multi-dimensional scaling plot (Fig. 1a). Dimension 1 showed the progression from the oldest to the newest cultivars. Genotypes were then chosen to be spread across year of registration as a cultivar to the UK market. Except for the first time-range which encompassed 130 years (1830–1959) of cultivar releases, all other ranges were split into decades and each time-range is represented by a similar number of genotypes (Fig. 1b). The final population was representative of breeding progress in cultivated barley for improved yield over time.
Selection of a two-row spring population. (a) Multidimensional scaling plot of 647 European two-row spring cultivars. Genotype information came from 2,336 previously published BOPA markers. The 209 selected cultivars forming the population in this study are shown as triangles. The year range represents the year each individual cultivar was registered. (b) Distribution of the selected population of 209 cultivars (orange) as part of the total 647 European two-row spring cultivars (green) by year of registration.
Pedigree data showed that modern barley germplasm is highly connected with most current genotypes’ descendants of a small number of “founder” genotypes (Pedigree file: Supplemental File 1, Pedigree attributes: Supplemental File 2). These supplemental pedigree files can be used as input for the pedigree visualisation tool Helium (https://helium.hutton.ac.uk/)62. Intermediate crosses were omitted from the file to be able to display the pedigree and produce a tree which is both readable and navigable. Using the pedigree data within Helium allows for further analyses.
Phenotyping
Field trials were done in 2019 and 2020 in three different locations: Minneapolis (Lat. 44.987, Long: −93.258; MN, USA), Dundee (Lat. 56.462, Long. −2.971; UK) and Gatersleben (Lat. 51.823, Long. 11.287; Germany). In total 29 agronomic traits were scored associated with development (earliness and growth habit traits), grain and height measurements. All the phenotypic data can be viewed and studied in a Germinate database: https://ics.hutton.ac.uk/germinate-barn/. Across years and sites, the results were consistent except for a few traits. All earliness traits showed a faster development to awn tipping all the way to peduncle senescence in Minnesota and slowest in Dundee. Outliers in the phenotypic scoring were the grain fertility measurements in Minnesota in 2019 where the spikes got stuck in the flag leaf sheath due to high temperatures during the growing season and did not emerge fully which led to a higher number of infertile florets. All phenotypic information was combined into BLUPs except for 24 out of 165 phenotype datasets which did not correlate with the rest (Supplemental Table 4 shows which phenotype values were combined; Supplemental Fig. 1 for distribution of BLUP values per phenotype). A strong positive correlation among the five different height measurements illustrate the robustness of the phenotypic dataset (Fig. 2).
Genotyping
To achieve the most extensive genotypic information for our population, variant calling from RNA-seq data, whole genome shotgun data and previously established 50 K SNP data was combined (32,484,981 raw SNPs). For RNA-seq and WGS, the data was filtered to keep only biallelic SNPs. We extracted the SNPs corresponding to the previous described BOPA markers across all 1463 sequencing datasets (six tissue-specific RNA-seq datasets with 209 genotypes each and one WGS dataset with 209 genotypes) for quality control. The Pearson correlation coefficient for all genotypes between datasets was calculated. This identified mixed-up samples where the genotype showed high correlation with a differently named sample and therefore allowed for correction of the genotypic information. Samples with a high number of heterogenous SNPs (above 10%) were removed as this pointed towards issues during sample preparation. The filtering step reduced the number of genotypes per tissue. The final numbers of genotypes per tissue varied between 191 to 199 (Supplemental Table 2 shows which genotypes per tissue where retained). The merged SNP file was filtered to remove highly heterozygous sites or those containing more than 20% missing data. The remaining sites were imputed using haplotype imputation. SNPs were pruned by LD using Plink to reduce the dataset size to the final 1,509,447 SNPs52. SNP distribution along the 7 chromosomes is shown in Fig. 3.
SNP and gene distribution along the seven barley chromosomes (a) SNP distribution and density of the final 1,509,447 SNPs in the genotypic marker file along the seven barley chromosomes in 1 Mb bins. The SNPs were identified from the 50k SNP array, RNA-sequencing and whole genome shotgun sequencing datasets and filtered to remove missing values and heterozygosity. (b) Gene density along the seven barley chromosomes.
Gene expression
RNA-seq data for six different tissues (crown, grain, inflorescence, peduncle, root, spikelet) was mapped against the BaRTv2 transcriptome using Salmon22. The expression of all 39,434 genes in transcript per million (TPM)55,56,57,58,59,60 for each tissue were used as input to generate a multidimensional scaling plot (MDS). The MDS shows all 209 genotypes cluster together by tissue type (Fig. 4). The tissue furthest separated by the first dimension from the rest was the root tissue. The two tissues sampled from the spikelet at green anther stages (spikelet) and developing grain at five days post anthesis (grain) show the highest overlap.
Data use-case scenarios
In the following three examples we show how the above datasets can be used.
In the first example the expression data has been used to filter for tissue-specific gene expression. Tissue-specific genes showed that the root tissue was the most distinct with 776 genes identified as root specific (Fig. 5). Overall, of the tissue specific genes, 572 genes were only expressed in grain, 437 in spikelet, 198 in inflorescence, 86 in peduncle and 64 in crown. Inflorescence and peduncle shared the highest overlap of expressed genes with 927 genes and 13,215 genes were expressed in all six tissues. While the MDS plot shows a high overlap of samples between spikelet and grain in the first two dimensions, the third dimension divides those tissues which fits with these two tissues showing the second and third highest tissue-specific gene expression. Gene ontology for the peduncle resulted in no significant terms. The Gene ontology results for all remaining five tissues are shown in Fig. 6. The associated terms were generally comparable to those previously identified in maize63.
Gene ontology (GO) enrichment for the tissue-specific genes in (a) root, (b) grain, (c) inflorescence, (d) spikelet and (e) crown. X-axis shows the percentage of genes associated with the GO term out of all genes in BaRTv2 associated with this term. Y-axis shows the significance as FDR adjusted -log(p-value) of the GO term. The area of the circle corresponds to the number of genes associated with the GO term.
In the second example, we illustrate how the data can be used to explore the potential impact of genetic variation on gene activity or protein function by identifying premature stop codons or frameshift mutations in a high confidence variant dataset. For the SNP dataset we started with 32 million SNPs, removed heterozygous SNPs and filtered for variants with less than 20% missing data and a minor allele frequency of 2.5% which resulted in 4,012,229 SNPs64. Those were used as input into SnpEff which identified 9,219,271 effects (as described by SnpEff: http://pcingola.github.io/SnpEff/se_inputoutput/#eff-field-vcf-output-files) caused by those 4 million SNPs. Of those effects, 4% (368,650) were in exons, with 53.78% synonymous variants, corresponding to 199,545 effects in 17,446 genes. The non-synonymous variants represented 45.57% (169,105 effects) of the exon effects in 19,057 genes and 0.65% classified as nonsense. The 0.65% corresponded to 2,425 transcripts and 1,105 genes with a premature stop codon in the sequence. For the Indel identification only the RNA-seq variant files were considered as those provided higher read depth for the genic regions. They were also filtered by removing heterozygous variants, keeping those with less than 20% missing data and a minor allele frequency of 2.5%. A total of 50,865 variants remained52 which SnpEff predicted to cause 558,991 effects. 50.31% (281,228 effects) were upstream or downstream of the gene and 41.81% (233,706 effects) in the intronic region. After filtering for disruptive frame shifts caused by insertions or deletions resulting in changes to the protein sequence, 1,912 genes remained which we designated as potentially non-functional in some of the cultivars64. Such structural variation can be explored in relation to gene expression. For example, Fig. 7 shows the expression of two genes BaRT2v18chr5HG260690 and BaRT2v18chr2HG058650 with frameshift mutations in comparison to the Barke reference allele. The consequence of all such observed variation still needs to be explored.
Gene expression in TPM (transcripts per million) for two genes identified with changes in the protein sequence. (a) BaRT2v18chr5HG260690 and (b) BaRT2v18chr2HG058650 split by haplotype on the x-axis. The first haplotype always represents the reference allele from the genotype Barke, and the second allele represents the alternative.
Third, we show a genome wide association study (GWAS) using the 1,509,447 SNP markers and the morphological character “awn length” as a phenotype. We used the Mixed Linear Model (MLM) in GAPIT47 to identify associations in the genome. Using a -log10(p) cut-off of 5 resulted in 6 significant peaks (Fig. 8). The most significant SNP was found on chromosome 5H at position 441 Mb within 1 kb of HvDep1 (BaRT2v18chr5HG247460) previously shown to influence awn length65. The other associations and traits remain to be explored.
Usage Notes
To perform the analysis using the Snakemake66 pipeline (see code availability) a high-performance computing (HPC) cluster is needed. For example, the Salmon indexing step in this setup needed 56 Gb of memory using 16 cores, mapping of each individual sample needed 31 Gb of memory using 8 cores. Downstream analyses like the genome wide association studies can be performed by downloading the BLUPs of the phenotypes and the marker file from Germinate.
Code availability
The code for analysing the RNA-sequencing data from mapping to genome and transcriptome to variant calling was combined into a Snakemake66 pipeline and is available on GitHub: https://github.com/SchreiberM/BARN.
References
FAO.FAOstat. License: CC BY-NC-SA 3.0 IGO. Extracted from: https://www.fao.org/faostat/. Data of Access: 16-12-2022, 2013).
O’Connor, A. Brewing and distilling in Scotland–economic facts and figures. Scottish Parliament Information Centre (2018).
Dawson, I. K. et al. Barley: a translational model for adaptation to climate change. New Phytol 206, 913–931, https://doi.org/10.1111/nph.13266 (2015).
Fischbeck, G. in Developments in Plant Genetics and Breeding Vol. 7 (eds von Bothmer, R., van Hintum, T., Knüpffer, H. & Sato, K.) 29–52 (Elsevier, 2003).
Ortiz, R., Nurminiemi, M., Madsen, S., Rognli, O. A. & Bjørnstad, Å. Genetic gains in Nordic spring barley breeding over sixty years. Euphytica 126, 283–289, https://doi.org/10.1023/A:1016302626527 (2002).
Schuster, W. H. Welchen Beitrag leistet die Pflanzenzüchtung zur Leistungssteigerung von Kulturpflanzenarten? Pflanzenbauwissenschaften 1, 9–18 (1997).
Gustafsson, A., Hagberg, A., Persson, G. & Wiklund, K. Induced mutations and barley improvement. Theor Appl Genet 41, 239–248, https://doi.org/10.1007/BF00277792 (1971).
Dockter, C. et al. Induced Variations in Brassinosteroid Genes Define Barley Height and Sturdiness, and Expand the Green Revolution Genetic Toolkit. Plant Physiology 166, 1912–1927, https://doi.org/10.1104/pp.114.250738 (2014).
Mammadov, J., Aggarwal, R., Buyyarapu, R. & Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. International Journal of Plant Genomics 2012, 728398, https://doi.org/10.1155/2012/728398 (2012).
Cockram, J. et al. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proceedings of the National Academy of Sciences of the United States of America 107, 21611–21616 (2010).
Comadran, J. et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nature Genetics 44, 1388–1392, https://doi.org/10.1038/ng.2447 (2012).
Matros, A. et al. Genome-wide association study reveals the genetic complexity of fructan accumulation patterns in barley grain. J Exp Bot 72, 2383–2402, https://doi.org/10.1093/jxb/erab002 (2021).
Ramsay, L. et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nature Genetics 43, 169, https://doi.org/10.1038/ng.745 (2011).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433, https://doi.org/10.1038/nature22043 (2017).
Jayakodi, M. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289, https://doi.org/10.1038/s41586-020-2947-8 (2020).
Bayer, M. M. et al. Development and Evaluation of a Barley 50k iSelect SNP Array. Front Plant Sci 8, 1792, https://doi.org/10.3389/fpls.2017.01792 (2017).
Tondelli, A. et al. Structural and Temporal Variation in Genetic Diversity of European Spring Two-Row Barley Cultivars and Association Mapping of Quantitative Traits. The Plant Genome 6, plantgenome2013.2003.0007, https://doi.org/10.3835/plantgenome2013.03.0007 (2013).
Looseley, M. E. et al. Association mapping of malting quality traits in UK spring and winter barley cultivar collections. Theor Appl Genet 133, 2567–2582, https://doi.org/10.1007/s00122-020-03618-9 (2020).
Thomas, W. et al. HGCA Project Report 528: Association genetics of UK elite barley (AGOUEB). (2014).
Russell, J. R. et al. A retrospective analysis of spring barley germplasm development from ‘foundation genotypes’ to currently successful cultivars. Molecular Breeding 6, 553–568, https://doi.org/10.1023/A:1011372312962 (2000).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14, 417–419, https://doi.org/10.1038/nmeth.4197 (2017).
Coulter, M. et al. BaRTv2: a highly resolved barley reference transcriptome for accurate transcript-specific RNA-seq quantification. Plant J https://doi.org/10.1111/tpj.15871 (2022).
Srivastava, A. et al. Alignment and mapping methodology influence transcript abundance estimation. Genome Biol 21, 239, https://doi.org/10.1186/s13059-020-02151-8 (2020).
R Core Team. (R Foundation for Statistical Computing, 2021).
Soneson, C., Love, M. & Robinson, M. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences [version 2; referees: 2 approved]. F1000Research 4, https://doi.org/10.12688/f1000research.7563.2 (2016).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, https://doi.org/10.1093/bioinformatics/btp616 (2010).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940, https://doi.org/10.1093/bioinformatics/btx364 (2017).
Gu, Z. Complex heatmap visualization. iMeta 1, e43, https://doi.org/10.1002/imt2.43 (2022).
Schreiber, M., Orr, J., Barakate, A. & Waugh, R. Barley (Hordeum Vulgare) Anther and Meiocyte RNA Sequencing: Mapping Sequencing Reads and Downstream Data Analyses. Methods Mol Biol 2484, 291–311, https://doi.org/10.1007/978-1-0716-2253-7_20 (2022).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, https://doi.org/10.1093/bioinformatics/bts635 (2013).
Broad Institute. Picard tools. Broad Institute, GitHub repository Version 2.18.4, http://broadinstitute.github.io/picard/ (2018).
Barnett, D. W., Garrison, E. K., Quinlan, A. R., Stromberg, M. P. & Marth, G. T. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692, https://doi.org/10.1093/bioinformatics/btr174 (2011).
Garrison, E. M. G. Haplotype-based variant detection from short-read sequencing. ArXiv e-prints, 9 (2012).
Milner, S. G. et al. Genebank genomics highlights the diversity of a global barley collection. Nat Genet 51, 319–326, https://doi.org/10.1038/s41588-018-0266-x (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3, https://doi.org/10.14806/ej.17.1.200 (2011).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100, https://doi.org/10.1093/bioinformatics/bty191 (2018).
Novocraft Technologies Sdn Bhd. Novosort. Version 3.06.05, https://www.novocraft.com/products/novosort/.
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, https://doi.org/10.1093/bioinformatics/btp352 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, https://doi.org/10.1093/gigascience/giab008 (2021).
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635, https://doi.org/10.1093/bioinformatics/btm308 (2007).
Swarts, K. et al. Novel Methods to Optimize Genotypic Imputation for Low-Coverage, Next-Generation Sequence Data in Crop Plants. The Plant Genome 7, plantgenome2014.2005.0023, https://doi.org/10.3835/plantgenome2014.05.0023 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, https://doi.org/10.1086/519795 (2007).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92, https://doi.org/10.4161/fly.19695 (2012).
Alvarado, G. et al. META-R: A software to analyze data from multi-environment plant breeding trials. The Crop Journal 8, 745–756, https://doi.org/10.1016/j.cj.2020.03.010 (2020).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208, https://doi.org/10.1038/ng1702 (2006).
Lipka, A. E. et al. GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399, https://doi.org/10.1093/bioinformatics/bts444 (2012).
ENA European Nucleotide Archive, https://identifiers.org/ena.embl:PRJEB49069 (2023).
ENA European Nucleotide Archive, https://identifiers.org/ena.embl:PRJEB48903 (2023).
Raubach, S. et al. From bits to bites: Advancement of the Germinate platform to support prebreeding informatics for crop wild relatives. Crop Science 61, 1538–1566, https://doi.org/10.1002/csc2.20248 (2021).
Cezard, T. et al. The European Variation Archive: a FAIR resource of genomic variation for all species. Nucleic Acids Res 50, D1216–D1220, https://doi.org/10.1093/nar/gkab960 (2022).
EVA European Variation Archive, https://identifiers.org/ebi/bioproject:PRJEB65875 (2023).
Arend, D. et al. e!DAL–a framework to store, share and publish research data. BMC Bioinformatics 15, 214, https://doi.org/10.1186/1471-2105-15-214 (2014).
Athar, A. et al. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res 47, D711–D715, https://doi.org/10.1093/nar/gky964 (2019).
Schreiber, M. RNA-seq data from spikelet tissues of cultivated two-row European spring barley genotypes. ArrayExpress https://doi.org/10.1101/2023.03.06.531259 (2023).
Schreiber, M. RNA-seq data from grain tissue of cultivated two-row European spring barley genotypes, ArrayExpress, https://identifiers.org/arrayexpress:E-MTAB-13236 (2023).
Schreiber, M. RNA-seq data from root tissue of cultivated two-row European spring barley genotypes, ArrayExpress, https://identifiers.org/arrayexpress:E-MTAB-13235 (2023).
Schreiber, M. RNA-seq data from crown tissue of cultivated two-row European spring barley genotypes, ArrayExpress, https://identifiers.org/arrayexpress:E-MTAB-13234 (2023).
Schreiber, M. RNA-seq data from inflorescence tissue of cultivated two-row European spring barley genotypes, ArrayExpress, https://identifiers.org/arrayexpress:E-MTAB-13233 (2023).
Schreiber, M. RNA-seq data from peduncle tissue of cultivated two-row European spring barley genotypes. ArrayExpress https://doi.org/10.1101/2023.03.06.531259 (2023).
Close, T. J. et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10, 582, https://doi.org/10.1186/1471-2164-10-582 (2009).
Shaw, P. D., Graham, M., Kennedy, J., Milne, I. & Marshall, D. F. Helium: visualization of large scale plant pedigrees. BMC Bioinformatics 15, 259, https://doi.org/10.1186/1471-2105-15-259 (2014).
Li, Y. et al. Genome-scale mining of root-preferential genes from maize and characterization of their promoter activity. BMC Plant Biol 19, 584, https://doi.org/10.1186/s12870-019-2198-8 (2019).
Schreiber, M. et al. Data record for the genomic resources of cultivated European two-rowed spring barley genotypes. e!Dal https://doi.org/10.5447/ipk/2023/15 (2023).
Wendt, T. et al. HvDep1 Is a Positive Regulator of Culm Elongation and Grain Size in Barley and Impacts Yield in an Environment-Dependent Manner. PLoS One 11, e0168924, https://doi.org/10.1371/journal.pone.0168924 (2016).
Molder, F. et al. Sustainable data analysis with Snakemake. F1000Res 10, 33, https://doi.org/10.12688/f1000research.29032.2 (2021).
Acknowledgements
The authors would like to acknowledge Richard Keith and Chris Warden for maintaining the JHI field trials, Nicola McCallum and Ruth Hamilton for the help with phenotypic scoring and John Fuller for the RNA-seq library preparation. We thank Micha Bayer, Runxuan Zhang, John Brown and Wenbin Guo for advice on the project design and bioinformatics. We would like to acknowledge Shane Heinen, Yadong Huang, Ismaél Pfeifer, and Letícia Pasqualino for the help in the lab and field work at the University of Minnesota. We acknowledge current and former members of the Genomics of Genetic Resources research group at IPK Gatersleben: Mary Ziems for planning and maintaining field trials, phenotypic scoring and tissue sampling; Mark Timothy Rabanus-Wallace, Hélène Pidon, Sudharsan Padmarasu, Mingjiu Li, Jayavardhan Reddy Kunam, Mohammed Rafaqat, Mohammad Awais, Jaqueline Pohl, Susanne König, Ines Walde, Manuela Knauft, Manuela Kretschmann and Beate Kamm for phenotypic scoring and tissue sampling. We thank Ines Walde and Susanne König at IPK for preparing sequencing libraries and generating sequencing data. Thanks are also given to the Research/Scientific Computing teams at The James Hutton Institute and NIAB for providing computational resources and technical support for the “UK’s Crop Diversity Bioinformatics HPC” (BBSRC grant BB/S019669/1), use of which has contributed to the results reported. The work was supported by funding from the Rural and Environment Science and Analytical Services Division of the Scottish Government. The authors acknowledge the Minnesota Supercomputing Institute at the University of Minnesota. The project received funding in frame of the ERA-CAPS Research Programme with funding (i) of the German partners through German Research Foundation (DFG) to NS under the project references MU 3589/1-1 | STE 1102/15-1 | WA 3336/4-1, (ii) the Scottish partner to RW through BBSRC (award number BB/S004610/1), (iii) the US partner to GJM through NSF (award number 1844331)
Author information
Authors and Affiliations
Contributions
M.S. wrote the manuscript, conducted data collection and analysis. R.Wo. conducted RNA-seq and W.G.S., data collection and analysis. AHa conducted RNA-seq data collection and analysis. M.C. did RNA-seq and data collection. J.R. provided seed material and did field trials. A.Hi. conducted RNA sequencing, whole genome shotgun sequencing and primary data analysis. AF did data management. G.J.M., N.S. and R.Wa. designed the experiment, supervised the work and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schreiber, M., Wonneberger, R., Haaning, A.M. et al. Genomic resources for a historical collection of cultivated two-row European spring barley genotypes. Sci Data 11, 66 (2024). https://doi.org/10.1038/s41597-023-02850-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02850-4