Abstract
The flatworm Schistosoma mansoni is an important but neglected pathogen that causes the disease schistosomiasis in millions of people worldwide. The parasite has a complex life cycle, undergoing sexual reproduction in a mammalian host and asexual replication in a snail host. Understanding the molecular mechanisms that the parasite uses to transition between hosts and develop into dimorphic reproductively competent adults may reveal new strategies for control. We present the first comprehensive transcriptomic analysis of S. mansoni, from eggs to sexually naïve worms. Focusing on eight life stages spanning free-living water-borne and parasitic stages from both intermediate and definitive hosts, we have generated deep RNA-seq data for five replicates per group for a total of 75 data sets. The data were produced using a single approach to increase the accuracy of stage-to-stage comparisons and made accessible via a user-friendly tool to visualise and explore gene expression (https://lifecycle.schisto.xyz/). These data are valuable for understanding the biology and sex-specific development of schistosomes and the interpretation of complementary genomic and functional genetics studies.
Similar content being viewed by others
Background & Summary
Schistosomiasis is a neglected tropical disease caused by parasitic flatworms of the genus Schistosoma (Phylum: Platyhelminthes, Class: Trematoda). The disease is primarily controlled via large-scale preventative chemotherapy programmes that distribute the anthelmintic drug praziquantel, and in 2021, over 75 million people in 51 countries received treatment1. Schistosoma mansoni is the most widespread and most studied schistosome species, which infects humans and causes intestinal schistosomiasis that is characterised by abdominal pain and diarrhoea, as well as hypertension in abdominal blood vessels, hepatosplenomegaly, liver granulomas, and fibrosis in chronic cases.
Schistosomes have a complex life cycle. Sexually mature, paired adult parasites excrete eggs in the faeces of their mammalian host that, upon water contact, hatch to release motile larvae called miracidia. The miracidia seek and infect freshwater intermediate host snails and undergo asexual clonal replication before re-entering the water as cercariae to infect the mammalian definitive host. Within the mammalian host, cercariae develop into schistosomula and traverse the vasculature until sexually naïve male and female parasites pair and mature, becoming reproductively competent in the mesenteric veins of the intestines (Fig. 1).
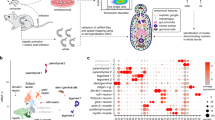
Sampling key stages of the Schistosoma mansoni life cycle. The flatworm S. mansoni undergoes a series of morphological transitions throughout development, using waterborne stages to transition between the intermediate snail host and the definitive mammalian host. The life stages and sex (male, female, mixed) collected for this study are shown with their corresponding sex symbols. Eggs are produced from paired, sexually mature adults; however, sexually reproducing adults were not sequenced in this study. Figure adapted from Laura Olivares Boldú/Wellcome Connecting Science.
Genomics approaches have provided great insight into the evolutionary history and life history traits of Schistosoma spp. For S. mansoni, this has been supported by ongoing efforts to develop high-quality reference genome resources2,3,4. As these genome resources have evolved, several studies have focused on characterising the transcriptome of various stages throughout the life cycle4,5,6,7,8,9,10, highlighting key genes involved in developmental transitions from free-living to parasitic stages and commitment towards sexual maturity. More recently, single-cell RNA sequencing on mammalian-infective stages11,12,13 and miracidia14 has been demonstrated, providing intricate details of body plans and cellular functions. However, a fundamental limitation of these existing transcriptomic data is that there is significant biological and technical variation across datasets, including but not limited to different parasite strains, numbers of replicates, and sequencing technologies and sampling depth, making it challenging to understand transcriptional changes across the entire life cycle or to compare data sets. Furthermore, most data sets have been sampled from pools of male and female parasites, limiting understanding of sex-specific transcriptomic changes across the life cycle.
Here, we present a comprehensive sex- and stage-specific RNA-seq data set to uncover transcriptional changes throughout the life cycle of S. mansoni. Our sampling (n = 75) of in vitro and in vivo cultured parasites includes five replicates each of mixed-sex eggs, miracidia, primary sporocysts, secondary sporocysts, cercariae, and schistosomules, as well as separate female and male secondary sporocysts, cercariae, schistosomules, and sexually naïve worms. Sexually reproducing worms were not chosen for sequencing in this study so that female-only data without egg contamination was sampled and because sexually-reproducing worms were the focus of previously published RNA-seq studies from this lab using the same methodology5,9,15.
To aid data exploration and reuse, we have implemented a simple tool – https://lifecycle.schisto.xyz/ – to visualise and subset transcript counts from these deep-coverage Illumina sequencing data. The data reported here will support future studies on understanding the sex-specific development of schistosomes and aid in interpreting functional studies describing gene function, drug-target discovery, and single-cell interpretation.
Methods
Ethics statement
All mouse procedures were conducted by authorised personnel under the Home Office Project Licence No. P77E8A062, held by Dr Gabriel Rinaldi (Wellcome Sanger Institute). All protocols were approved by the Animal Welfare and Ethical Review Body (AWERB) of the Wellcome Sanger Institute as required by the UK Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012.
Collection of parasite samples
The complete S. mansoni life cycle was maintained at the Wellcome Sanger Institute, as previously described16. The S. mansoni NMRI stain (Puerto Rican) was used to infect female outbred TO-1 mice between 8–12 weeks old and the Biomphalaria glabrata NMRI strain (received from Prof M. Doenhoff). Individual life stages were collected as follows:
Eggs
Patently infected mice at 42 days post-exposure were culled using an overdose of pentobarbitone containing 10 U/ml heparin. The livers were dissected and washed in 1x PBS with 2% antibiotic-antimycotic and 70% ethanol. The mouse livers were minced using a sterile disposable scalpel, enzymatically degraded with collagenase, and passed through a 250 μm, followed by a 150 µm, sieve. Schistosome eggs were purified using density centrifugation through a sucrose-Percoll gradient17,18. Eggs from a pool of five livers of mice exposed to the same mixed-sex collection of cercariae represent one biological replicate.
Miracidia
Eggs from five livers for each replicate, prepared as above, were hatched under direct light at ~28 °C in MilliQ water in 24-well plates19,20. Miracidia were collected every 30 minutes for three hours.
Primary (mother) sporocysts (1-day & 5-day; in vitro)
Miracidia were transformed in vitro by centrifugation and then resuspended in sporocyst culture medium, which was maintained at 28 °C under malaria gas (90–92% N, 5% CO2, and 3–5% O2) with daily media changes21,22,23.
Secondary (daughter) sporocysts (32-day; snail)
For mixed-sex infections, individual B. glabrata snails were each exposed to 15 miracidia. For single-sex infections, each B. glabrata snail was exposed to a single miracidium. Each biological replicate represents an infection initiated from a different miracidia pool. Clonal, asexual reproduction of the parasite within the snail host results in a single-sex infection, and the cercariae developed from mono miracidium-infected snails are all single-sex. At 32 days post-exposure to miracidia/miracidium, B. glabrata snails were dissected, and secondary sporocysts (also known as daughter sporocysts) were collected with particular attention to removing all visible snail tissue.
Cercariae
After ~4–8 weeks post-exposure to miracidia/miracidium, snails begin to release cercariae into the water. Cercariae were released by exposing patent snails to bright light at 28 °C for up to 2 hours.
Schistosomules
Mixed-sex or single-sex cercariae were shed from snails, as above, and placed on ice for 30 mins. Cercariae tails were removed by repeated centrifugation and resuspension in 1x PBS with 2% antibiotic-antimycotic. The parasites were placed in schistosomula medium at 37 °C and 5% CO2 overnight. The following day, the separated tails were removed with a transfer pipet. Transformed schistosomula were maintained in vitro at 37 °C and 5% CO2 with daily media changes20,24,25,26,27.
Sexually naïve juvenile worms
Mice were percutaneously infected with 250 mixed-sex cercariae via the tail, as previously described16. At 26 days post-exposure, sexually naïve worms were recovered via portal perfusion with approximately 30 ml of Dulbecco’s Modified Eagle Medium (DMEM; high glucose) and 10 U/ml heparin20,28.
After collection, all stages were gently centrifuged, washed with 1x PBS, and then stored at −80 °C in 500 µl of TRIzol Reagent (Invitrogen) until RNA extraction.
Discrimination of sex of early monomorphic life stages
Early parasite life stages are sexually monomorphic and cannot be differentiated morphologically until the sexually naïve adult stages. To identify both male and female stages from 32-day sporocysts (after clonal amplification) onward, we determined the sex of shedding cercariae from snails29,30 after mono-miracidium infections using a modified HOTSHOT method (https://health.uconn.edu/mouse-genome-modification/protocols/hotshot-method-of-dna-preparation/) to obtain DNA followed by PCR-amplification of the female-specific W1 repeat31,32,33. Snails shedding single-sex parasites were then used to generate sex-defined parasite material for subsequent stages.
RNA isolation, library preparation, and sequencing
In preparation for RNA isolation, samples were thawed on ice, transferred to MagNA Lyser Green Beads (Roche) tubes, and homogenised using the FastPrep-24 instrument (MP Biomedical) for 2 × 30 secs. An additional 500 µl of TRIzol was added to each sample, and the homogenate was allowed to sit at room temperature for five minutes to complete the dissociation of nucleoprotein complexes. RNA was isolated using TRIzol Reagent (Invitrogen) manufacturer’s instructions.
After isolation, RNA was immediately cleaned and concentrated using the RNA Clean and Concentrator-5 kit (Zymo Research) with on-column DNase I treatment according to the manufacturer’s instructions. Total RNA was quantified on the Qubit 3 Fluorometer (Invitrogen), and quality was analysed on the 2100 Bioanalyzer with RNA 6000 Nano-chips (Agilent Technologies). RNA was stored at −80 °C until library preparation.
RNA-seq libraries were prepared with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Sequencing libraries were quantified by qPCR, and their concentration was normalised before equimolar pooling. Libraries were sequenced using 150 bp paired-end read chemistry on two S4 lanes of an Illumina NovaSeq6000 platform.
RNA-seq data analysis
Demultiplexed FASTQ reads were trimmed using TrimGalore v0.4.4 (https://github.com/FelixKrueger/TrimGalore). Trimmed sequencing reads were mapped to the S. mansoni reference genome v103 from WormBase ParaSite v18 (https://parasite.wormbase.org/Schistosoma_mansoni_prjea36577/Info/Index/34) using STAR v2.7.9a35 (parameters:–alignIntronMin 10–outSAMtype BAM SortedByCoordinate–limitBAMsortRAM 90000000000). The S. mansoni v10 annotation (available from WormBase ParaSite v18) was used to guide read mapping; the genome annotation GFF3 file was converted to a GTF file using gffread from the cufflinks (v2.2.1) package36. Mapped read sets belonging to the same sample were combined using samtools (v1.9) merge. Protein-coding features were quantified from the results of STAR using StringTie v2.1.435.
Principal component analysis (PCA) was performed in R (version 4.1.3) using the prcomp() command and the log2-transformed transcript per million (TPM) count matrix. To generate the heatmap, pairwise sample distances (type = euclidean), using TMP values of the top 25% most variable genes, were calculated in R using the dist() package.
RNA-seq data visualisation
We used Seurat v4.2.1 (https://satijalab.org/seurat/37) to construct an object using the log2-transformed TPM count matrix (log2(TPM + 1)). Metadata, including sex, stages, and sample identities, were added accordingly. To view the data in a lower dimensionality space, the top 2000 variable features were extracted using the FindVariableFeatures() function and data were then scaled for PCA analysis. The resulting Seurat object with 2D PCA embeddings, log2(TPM + 1) data, and metadata information was deployed using the interactive visualisation tool Cirrocumulus (https://github.com/lilab-bcb/cirrocumulus) with customisations and can be explored at https://lifecycle.schisto.xyz. The data files used to generate this website are available in the GitHub repository https://github.com/SKBuddenborg/Smansoni_rnaseq and are archived at Zenodo (https://doi.org/10.5281/zenodo.7849283)38, allowing researchers to recreate the visualisations, customise, and add data sets.
Data Records
The data set consists of bulk RNA-seq data from 75 samples, representing eight life stages, each with five replicates. For egg, miracidia, and 1-day and 5-day sporocysts, samples were mixed sex. For 32-day sporocysts, cercariae, 2-day schistosomules and 26-day sexually-naïve worms, pooled male, pooled female, and mixed-sex stages were collected. Sample metadata, including read counts, mapped reads, and ENA accession information, are described in Supplementary Table 1. All sequence data can be obtained from the European Nucleotide Archive (ENA) under project accession PRJEB3283939. The S. mansoni v10 genome and annotation used in the analysis are available from WormBase ParaSite release 18 (https://parasite.wormbase.org/Schistosoma_mansoni_prjea36577/Info/Index/).
Technical Validation
The dataset consists of approximately 4.4 × 109 sequencing reads, with an average of 58.9 million reads per sample (range: 21.3–183.3 million reads). Per sample, on average, 67.4% of reads mapped to the v10 assembly of the S. mansoni; for most life stages (excluding 32-day sporocysts), this percentage was higher (average: 77.1%); however, 32-day sporocysts showed a considerably lower percentage of mapped reads (average: 28.1%), which can be attributed to contamination from the snail host.
To assess the replicability of the data, we calculated TPM values per gene for each sample, which we analysed by principal component analysis (PCA; Fig. 2a) and clustering of Euclidean distance (Fig. 2b). PCA, which explained 70% of the variance in the first two principal components, showed clear clustering of replicate samples with only minor overlap between some life stages; however, sex differences within a life stage were less discernible. Differentiating stage and sex was more apparent based on the clustering of TPM by Euclidean distance, with clear discrimination of cercariae, sexually naïve worms, and eggs. Two-day schistosomules and 32-day sporocysts were also clearly differentiated by life stage; however, each contained some misclustering based on sex (single replicate for mixed sex/female for 2-day schistosomules; two replicates for male/female samples 32-day sporocysts), emphasising the importance of our relatively high number of biological replicates. Similarly, there was some misclustering between a single replicate of miracidia and 1-day sporocysts and a single 1-day and 5-day sporocyst; however, both cases represent transitions between adjacent stages in the life cycle. Overall, most replicates were clustered with their corresponding sex- and stage-specific data sets.
Clustering of RNA-seq data by parasite sex and life stage. (a) A PCA plot of the 25% most variable genes was calculated by the coefficient of variation, highlighting life stage (colour) and sex (shape). (b) Clustering of samples based on Euclidean distance (Coefficient of Variation; CoV) of the 25% most variable genes. In both panels a and b, the colour is consistent with Fig. 1.
Usage Notes
These data provide a rich resource to understand the sex and stage-specific developmental changes of S. mansoni throughout its life cycle. We have implemented a simple tool to visualise and subset these data to aid data exploration and reuse: https://lifecycle.schisto.xyz/. To illustrate, we highlight variation in the expression of S. mansoni Kunitz protease inhibitors that are proposed to be involved in the defence mechanisms of the parasite within the mammalian host40; in the v5 genome assembly, only a single copy was identified, whereas in the v10 genome assembly, 11 copies were identified3. Here, we demonstrate two approaches to visualise samples based on sex (Fig. 3a) and life stage (Fig. 3b), and differences in gene expression, either by colouring the PCA based on levels of gene expression represented as log2(TPM + 1) (Fig. 3c) or by displaying mean log2(TPM + 1) (combining each life stage) as a dotplot heatmap (Fig. 3d). Further customisation (colours, plots) and filtering (min, max expression, conditions) of the data are possible depending on the user’s preference to explore gene expression of any gene present in the genome. These analyses identify four groups of genes with distinct stage-specific expression patterns.
Visualisation and exploration of stage and sex-specific RNA-seq data of Schistosoma mansoni. Demonstration of our easy-to-use data exploration tool https://lifecycle.schisto.xyz/ using S. mansoni Kunitz protease inhibitors as an example. (a,b) PCA plot of RNA-seq data coloured by life stage and parasite sex, which highlights male, female, and mixed-sex parasite data to provide context for gene-specific analyses. (c) A series of PCA plots show the expression of 11 S. mansoni Kunitz protease inhibitors. In panels a, b, and c, each point represents a single replicate (n = 75) and is coloured by log transcript per million values (log2[TPM + 1]), with low expression represented by dark and high expression light colour. (d) A Heatmap of mean log2(TPM + 1) expression for each gene is presented in panel c. Each point represents the mean TMP from five replicates, with the size of the point proportional to the number of replicates within a group with data.
Code availability
All code used to analyse and visualise this data set has been deposited in the GitHub repository https://github.com/SKBuddenborg/Smansoni_rnaseq and is archived at Zenodo (https://doi.org/10.5281/zenodo.7849283)38.
References
World Health Organization. Schistosomiasis and soil-transmitted helminthiases: progress report, 2021. https://www.who.int/publications/i/item/who-wer9748-621-632 (2022).
Berriman, M. et al. The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358 (2009).
Buddenborg, S. K. et al. Assembled chromosomes of the blood fluke Schistosoma mansoni provide insight into the evolution of its ZW sex-determination system. bioRxiv 2021.08.13.456314 https://doi.org/10.1101/2021.08.13.456314 (2021).
Protasio, A. V. et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 6, e1455 (2012).
Wangwiwatsin, A. et al. Transcriptome of the parasitic flatworm Schistosoma mansoni during intra-mammalian development. PLoS Negl. Trop. Dis. 14, e0007743 (2020).
Wang, B., Collins, J. J. 3rd & Newmark, P. A. Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. Elife 2, e00768 (2013).
Anderson, L. et al. Schistosoma mansoni Egg, Adult Male and Female Comparative Gene Expression Analysis and Identification of Novel Genes by RNA-Seq. PLoS Negl. Trop. Dis. 9, e0004334 (2015).
Picard, M. A. L. et al. Sex-Biased Transcriptome of Schistosoma mansoni: Host-Parasite Interaction, Genetic Determinants and Epigenetic Regulators Are Associated with Sexual Differentiation. PLoS Negl. Trop. Dis. 10, e0004930 (2016).
Lu, Z. et al. A gene expression atlas of adult Schistosoma mansoni and their gonads. Sci Data 4, 170118 (2017).
Protasio, A. V. et al. MiR-277/4989 regulate transcriptional landscape during juvenile to adult transition in the parasitic helminth Schistosoma mansoni. PLoS Negl. Trop. Dis. 11, e0005559 (2017).
Diaz Soria, C. L. et al. Single-cell atlas of the first intra-mammalian developmental stage of the human parasite Schistosoma mansoni. Nat. Commun. 11, 6411 (2020).
Wendt, G. et al. A single-cell RNA-seq atlas of Schistosoma mansoni identifies a key regulator of blood feeding. Science 369, 1644–1649 (2020).
Li, P. et al. Single-cell analysis of Schistosoma mansoni identifies a conserved genetic program controlling germline stem cell fate. Nat. Commun. 12, 485 (2021).
Attenborough, T. et al. A single-cell atlas of the free-living miracidium larva of Schistosoma mansoni. bioRxiv 2023.03.27.533868 https://doi.org/10.1101/2023.03.27.533868 (2023).
Lu, Z. et al. Schistosome sex matters: a deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci. Rep. 6, 31150 (2016).
Crosnier, C. et al. Systematic screening of 96 cell-surface and secreted antigens does not identify any strongly protective vaccine candidates in a mouse model of infection. Wellcome Open Res 4, 159 (2019).
Dalton, J. P., Day, S. R., Drew, A. C. & Brindley, P. J. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology 115, 29–32 (1997).
Mann, V. H., Morales, M. E., Rinaldi, G. & Brindley, P. J. Culture for genetic manipulation of developmental stages of Schistosoma mansoni. Parasitology 137, 451–462 (2010).
Lewis, F. A., Stirewalt, M. A., Souza, C. P. & Gazzinelli, G. Large-scale laboratory maintenance of Schistosoma mansoni, with observations on three schistosome/snail host combinations. J. Parasitol. 72, 813–829 (1986).
Tucker, M. S., Karunaratne, L. B., Lewis, F. A., Freitas, T. C. & Liang, Y.-S. Schistosomiasis. Curr. Protoc. Immunol. 103, 19.1.1–19.1.58 (2013).
Ivanchenko, M. G. et al. Continuous in vitro propagation and differentiation of cultures of the intramolluscan stages of the human parasite Schistosoma mansoni. Proc. Natl. Acad. Sci. USA 96, 4965–4970 (1999).
Buddenborg, S. K. Isolation of Schistosoma mansoni eggs, miracidia, and sporocysts for in vitro cultivation. Protocols.io. https://doi.org/10.17504/protocols.io.81wgbye5yvpk/v1 (2023).
Bixler, L. M. et al. Axenic culture of Schistosoma mansoni sporocysts in low O2 environments. J. Parasitol. 87, 1167–1168 (2001).
Colley, D. G. & Wikel, S. K. Schistosoma mansoni: simplified method for the production of schistosomules. Exp. Parasitol. 35, 44–51 (1974).
Cousin, C. E., Stirewalt, M. A. & Dorsey, C. H. Schistosoma mansoni: ultrastructure of early transformation of skin- and shear-pressure-derived schistosomules. Exp. Parasitol. 51, 341–365 (1981).
Lazdins, J. K., Stein, M. J., David, J. R. & Sher, A. Schistosoma mansoni: rapid isolation and purification of schistosomula of different developmental stages by centrifugation on discontinuous density gradients of Percoll. Exp. Parasitol. 53, 39–44 (1982).
Buddenborg, S. K. Schistosoma mansoni cercariae transformation (without needle). Protocols.io. https://doi.org/10.17504/protocols.io.8epv5jp36l1b/v2 (2023).
Duvall, R. H. & DeWitt, W. B. An improved perfusion technique for recovering adult schistosomes from laboratory animals. Am. J. Trop. Med. Hyg. 16, 483–486 (1967).
Lewis, F. Schistosomiasis. Curr. Protoc. Immunol. 19.1, (2001).
Buddenborg, SK., Sankaranarayanan, G., Lotkowska, ME., McCarthy, C., Wangwiwatsin, W., Dias Soria, CL., Seymour, L., Rinaldi, G. & Berriman, M. Schistosoma mansonicercariae shedding, Protocols.io., https://doi.org/10.17504/protocols.io.eq2ly7bbplx9/v1 (2023).
Buddenborg, SK. & Berriman, M. Schistosoma mansonicercariae sexing, Protocols.io., https://doi.org/10.17504/protocols.io.rm7vzby12vx1/v1 (2022).
Webster, P., Mansour, T. E. & Bieber, D. Isolation of a female-specific, highly repeated Schistosoma mansoni DNA probe and its use in an assay of cercarial sex. Mol. Biochem. Parasitol. 36, 217–222 (1989).
Gasser, R. B., Morahan, G. & Mitchell, G. F. Sexing single larval stages of Schistosoma mansoni by polymerase chain reaction. Mol. Biochem. Parasitol. 47, 255–258 (1991).
Howe, K. L., Bolt, B. J., Shafie, M., Kersey, P. & Berriman, M. WormBase ParaSite − a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2–10 (2017).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Buddenborg, S. K. Dataset: Schistosoma mansoni sex and stage specific RNAseq: metadata, transcript counts, code and figures. Zenodo. https://doi.org/10.5281/zenodo.7849283 (2023).
Project PRJEB32839. The_stage_and_sex_specific_transcriptome_of_the_human_parasite_Schistosoma_mansoni. European Nucleotide Archive https://identifiers.org/ena.embl:PRJEB32839 (2023).
Ranasinghe, S. L., Fischer, K., Gobert, G. N. & McManus, D. P. Functional expression of a novel Kunitz type protease inhibitor from the human blood fluke Schistosoma mansoni. Parasit. Vectors 8, 408 (2015).
Acknowledgements
We thank the Pathogen Informatics group at the Wellcome Sanger Institute for informatics support. We also thank Gabriel Rinaldi and other members of the Berriman Lab for discussions and support with the maintenance of the S. mansoni life cycle. This work was supported by the Wellcome Trust through core funding to the Wellcome Sanger Institute [206194]. SRD is supported by a UKRI Future Leaders Fellowship [MR/T020733/1]. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
S.K.B. and M.B. conceived and designed the study, S.K.B. and G.S. performed the experiments, S.K.B. analysed RNA-seq data, Z.L. supported the data visualisation, S.R.D. supervised data analysis and wrote the manuscript with S.K.B., with contributions from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buddenborg, S.K., Lu, Z., Sankaranarayan, G. et al. The stage- and sex-specific transcriptome of the human parasite Schistosoma mansoni. Sci Data 10, 775 (2023). https://doi.org/10.1038/s41597-023-02674-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02674-2