Abstract
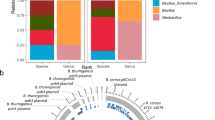
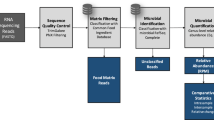
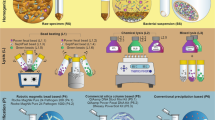
Deep investigation of the microbiome of food-production and food-processing environments through whole-metagenome sequencing (WMS) can provide detailed information on the taxonomic composition and functional potential of the microbial communities that inhabit them, with huge potential benefits for environmental monitoring programs. However, certain technical challenges jeopardize the application of WMS technologies with this aim, with the most relevant one being the recovery of a sufficient amount of DNA from the frequently low-biomass samples collected from the equipment, tools and surfaces of food-processing plants. Here, we present the first complete workflow, with optimized DNA-purification methodology, to obtain high-quality WMS sequencing results from samples taken from food-production and food-processing environments and reconstruct metagenome assembled genomes (MAGs). The protocol can yield DNA loads >10 ng in >98% of samples and >500 ng in 57.1% of samples and allows the collection of, on average, 12.2 MAGs per sample (with up to 62 MAGs in a single sample) in ~1 week, including both laboratory and computational work. This markedly improves on results previously obtained in studies performing WMS of processing environments and using other protocols not specifically developed to sequence these types of sample, in which <2 MAGs per sample were obtained. The full protocol has been developed and applied in the framework of the European Union project MASTER (Microbiome applications for sustainable food systems through technologies and enterprise) in 114 food-processing facilities from different production sectors.
Key points
-
This protocol outlines a procedure for sampling the microbiomes of environments with low-biomass yields such as those in a clean food-processing facility and analyzing them through WMS.
-
The procedure includes an optimized DNA-extraction stage to maximize DNA yield and allow WMS-based analysis, offering a more complete analysis of the microbiome than targeted methods currently used in industry and avoiding issues of bias associated with targeted high-throughput sequencing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw reads are available on the Sequence Read Archive of the NCBI under the BioProjects numbers PRJNA897099 (for vegetable facilities), PRJNA941197 (for an ice-cream facility), PRJNA997800 (for meat facilities), PRJNA997821 (for cheese facilities, except those located in Ireland) and PRJNA996188 for control samples. Raw reads for fish-processing factories and Irish cheese factories are available on the European Nucleotide Archive database under the accession numbers PRJEB62794 and PRJEB63604, respectively.
Code availability
The code used for raw reads filtering, assembly and binning into MAGs is available at https://github.com/SegataLab/MASTER-WP5-pipelines.
References
Møretrø, T. & Langsrud, S. Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Saf. 16, 1022–1041 (2017).
De Filippis, F., Valentino, V., Alvarez-Ordóñez, A., Cotter, P. D. & Ercolini, D. Environmental microbiome mapping as a strategy to improve quality and safety in the food industry. Curr. Opin. Food Sci. 38, 168–176 (2021).
Alvarez-Ordóñez, A., Coughlan, L. M., Briandet, R. & Cotter, P. D. Biofilms in food processing environments: challenges and opportunities. Annu. Rev. Food Sci. Technol. 10, 173–195 (2019).
Fagerlund, A., Langsrud, S. & Møretrø, T. Microbial diversity and ecology of biofilms in food industry environments associated with Listeria monocytogenes persistence. Curr. Opin. Food Sci. 37, 171–178 (2021).
Koutsoumanis, K. et al. Whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food‐borne microorganisms. EFSA J. 17, e05898 (2019).
Yap, M. et al. Next-generation food research: use of meta-omic approaches for characterizing microbial communities along the food chain. Annu. Rev. Food Sci. Technol. 13, 361–384 (2022).
Capouya, R. D., Mitchell, T., Clark, D. I., Clark, D. L. & Bass, P. D. A survey of microbial communities on dry-aged beef in commercial meat processing facilities. Meat Muscle Biol. 4, 1–11 (2020).
Zwirztz, B. et al. The sources and transmission routes of microbial populations throughout a meat processing facility. NPJ Biofilms Microbiomes 6, 26 (2020).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Xie, F. et al. An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 9, 137 (2021).
Olm, M. R. et al. Genome-resolved metagenomics of eukaryotic populations during early colonization of premature infants and in hospital rooms. Microbiome 7, 26 (2019).
Lind, A. L. & Pollard, K. S. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome 9, 58 (2021).
Santos-Medellin, C. et al. Viromes outperform total metagenomes in revealing the spatiotemporal patterns of agricultural soil viral communities. ISME J. 15, 1956–1970 (2021).
Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 6, 960–970 (2021).
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017).
Kim, H., Kim, M., Kim, S., Lee, Y. M. & Shin, S. C. Characterization of antimicrobial resistance genes and virulence factor genes in an Arctic permafrost region revealed by metagenomics. Environ. Pollut. 294, 118634 (2022).
Picone, N. et al. Metagenome assembled genome of a novel verrucomicrobial methanotroph from Pantelleria Island. Front. Microbiol. 12, 666929 (2021).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Yilmaz, S., Allgaier, M. & Hugenholtz, P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods 7, 943–944 (2010).
Valentino, V. et al. Evidence of virulence and antibiotic resistance genes from the microbiome mapping in minimally processed vegetables producing facilities. Food Res. Int. 162, 112202 (2022).
McHugh, A. J. et al. Microbiome-based environmental monitoring of a dairy processing facility highlights the challenges associated with low microbial-load samples. NPJ Sci. Food 5, 4 (2021).
Cobo-Díaz, J. F. et al. Microbial colonization and resistome dynamics in food processing environments of a newly opened pork cutting industry during 1.5 years of activity. Microbiome 9, 204 (2021).
Bruggeling, C. E. et al. Optimized bacterial DNA isolation method for microbiome analysis of human tissues. Microbiologyopen 10, e1191 (2021).
Hinlo, R., Gleeson, D., Lintermans, M. & Furlan, E. Methods to maximise recovery of environmental DNA from water samples. PLoS ONE 12, e0179251 (2017).
Yap, M., O’Sullivan, O., O’Toole, P. W. & Cotter, P. D. Development of sequencing-based methodologies to distinguish viable from non-viable cells in a bovine milk matrix: a pilot study. Front. Microbiol. 13, 1036643 (2022).
Chng, K. R. et al. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 26, 941–951 (2020).
Echeverría-Beirute, F., Varela-Benavides, I., Jiménez-Madrigal, J. P., Carvajal-Chacon, M. & Guzmán-Hernández, T. eDNA extraction protocol for metagenomic studies in tropical soils. Biotechniques 71, 580–586 (2021).
Jiang, W. et al. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 10, 768–779 (2015).
Fadiji, A. E. & Babalola, O. O. Metagenomics methods for the study of plant-associated microbial communities: a review. J. Microbiol. Methods 170, 105860 (2020).
Cabello-Yeves, P. J. et al. The microbiome of the Black Sea water column analyzed by shotgun and genome centric metagenomics. Environ. Microbiome 16, 5 (2021).
Keeratipibul, S. et al. Effect of swabbing techniques on the efficiency of bacterial recovery from food contact surfaces. Food Control 77, 139–144 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Karstens, L. et al. Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 4, 2379–5077 (2019).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023).
Acknowledgements
This work was funded by the European Commission under the European Union’s Horizon 2020 research and innovation program under grant agreement no. 818368 (MASTER). C.B. is grateful to Junta de Castilla y León and the European Social Fund for awarding her a pre-doctoral grant (BOCYL-D-07072020-6). A.P. is grateful to Ministerio de Ciencia e Innovación for awarding her a pre-doctoral grant (PRE2021-098910). N.M.Q. is currently funded by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 101034371. We thank AV Star Systems for their role in creating the Supplementary Video, and M. Coakley and S. Mortensen for their help in its preparation.
Author information
Authors and Affiliations
Contributions
M.L., M.P., D.O’N., V.T.M., M.W., A.M., N.S., P.D.C., D.E. and A.A.-O. conceived the study and obtained the funding. J.F.C.-D., C.B., F.D.F., V.V., R.C.R., I.C.-T., C.S., S.D., P.R.-M., N.M.Q., M.D., S.S., S.K. and A.P. performed the samplings at food-processing facilities. D.O’N. and L.M.d.S. designed and tested the improvements in the DNA-extraction protocol, and C.B., F.D.F., V.V., R.C.R. and A.P. tested the different versions of the DNA-extraction protocol for optimization. C.B., F.D.F., R.C.R., I.C.T., C.S., S.D., P.R.-M., N.M.Q., M.D., S.S., S.K. and A.P. applied the improved DNA-extraction protocol on samples from the food industry. F.A., F.P. and N.S. sequenced the extracted DNA. N.C., A.B.-M. and F.P. performed the bioinformatic analyses. J.F.C.-D., F.D.F., V.V., R.C.R., N.C., C.S. and N.M.Q. collated all the information. L.M.d.S., J.F.C.D. and C.B. prepared the figures. J.F.C.D., C.B. and A.A.-O. wrote the manuscript with input from all the authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
D.O’N. and L.M.d.S. are employees of QIAGEN GmbH. All other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Lena Florl, Andrea Moreno Switt, Bernard Taminiau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Valentino, V. et al. Food Res. Int. 162, 112202 (2022): https://doi.org/10.1016/j.foodres.2022.112202
Supplementary information
Supplementary Information
Supplementary Fig. 1, Note and Methods
Supplementary Video 1
Microbiome mapping in the food industry: a detailed visual procedure on how to prepare the materials and take the samples at a food-processing facility. The steps that should be followed in the laboratory for sample pre-processing are shown.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Barcenilla, C., Cobo-Díaz, J.F., De Filippis, F. et al. Improved sampling and DNA extraction procedures for microbiome analysis in food-processing environments. Nat Protoc (2024). https://doi.org/10.1038/s41596-023-00949-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-023-00949-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.