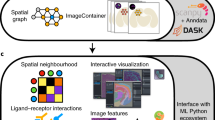
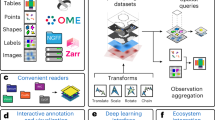
Abstract
Advances in spatial omics technologies have improved the understanding of cellular organization in tissues, leading to the generation of complex and heterogeneous data and prompting the development of specialized tools for managing, loading and visualizing spatial omics data. The Spatial Omics Database (SODB) was established to offer a unified format for data storage and interactive visualization modules. Here we detail the use of Pysodb, a Python-based tool designed to enable the efficient exploration and loading of spatial datasets from SODB within a Python environment. We present seven case studies using Pysodb, detailing the interaction with various computational methods, ensuring reproducibility of experimental data and facilitating the integration of new data and alternative applications in SODB. The approach offers a reference for method developers by outlining label and metadata availability in representative spatial data that can be loaded by Pysodb. The tool is supplemented by a website (https://protocols-pysodb.readthedocs.io/) with detailed information for benchmarking analysis, and allows method developers to focus on computational models by facilitating data processing. This protocol is designed for researchers with limited experience in computational biology. Depending on the dataset complexity, the protocol typically requires ~12 h to complete.
Key points
-
Pysodb allows researchers to load and explore spatial omics data in a Python environment. Data loaded using Pysodb follow the AnnData format, thus providing a unified format for storing over 3,000 datasets and facilitating benchmarking and reuse of data.
-
Alternative packages such as Scanpy, Squidpy and Giotto focus on data analysis; Pysodb complements them by providing a support platform for data storage and handling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The spatial datasets discussed in this protocol are available from SODB (https://gene.ai.tencent.com/SpatialOmics/). We provide guidelines on how to load and visualize these data at https://protocols-pysodb.readthedocs.io/en/latest/SOView/SOView.html#. The mouse cortex single-cell data are provided at https://figshare.com/articles/dataset/Visium/22332667. The human PDAC single-cell data are provided at https://figshare.com/articles/dataset/PDAC/22332574.
Code availability
Pysodb is a freely available software package written in the Python programming language. Source code can be found at https://github.com/TencentAILabHealthcare/pysodb. Installation instructions can be found at https://pysodb.readthedocs.io/en/latest/. The code used in this paper can be found at https://protocols-sodb.readthedocs.io/en/latest/. A Python version of SOView code can be found at https://github.com/yuanzhiyuan/SOView. An SOView tutorial can be found at https://soview-doc.readthedocs.io/en/latest/index.html.
References
Moffitt, J. R., Lundberg, E. & Heyn, H. The emerging landscape of spatial profiling technologies. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00515-3 (2022).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet.https://doi.org/10.1038/s41576-023-00580-2 (2023).
Rao, A., Barkley, D., Franca, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-01182-1 (2022).
Andreou, C., Weissleder, R. & Kircher, M. F. Multiplexed imaging in oncology. Nat. Biomed. Eng. 6, 527–540 (2022).
Hildebrandt, F. et al. Spatial transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat. Commun. 12, 7046 (2021).
Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell https://doi.org/10.1016/j.cell.2022.04.003 (2022).
Lohoff, T. et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-01006-2 (2021).
Marshall, J. L. et al. High-resolution Slide-seqV2 spatial transcriptomics enables discovery of disease-specific cell neighborhoods and pathways. iScience 25, 104097 (2022).
Yuan, Z. et al. SEAM is a spatial single nuclear metabolomics method for dissecting tissue microenvironment. Nat. Methods 18, 1223–1232 (2021).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 (2018).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell https://doi.org/10.1016/j.cell.2020.07.005 (2020).
Li, Y. et al. SOAR: a spatial transcriptomics analysis resource to model spatial variability and cell type interactions. Preprint at bioRxiv https://doi.org/10.1101/2022.04.17.488596 (2022).
Fan, Z., Chen, R. & Chen, X. SpatialDB: a database for spatially resolved transcriptomes. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz934 (2019).
Xu, Z. et al. STOmicsDB: a comprehensive database for spatial transcriptomics data sharing, analysis and visualization. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad933 (2023).
Yuan, Z. et al. SODB facilitates comprehensive exploration of spatial omics data. Nat. Methods https://doi.org/10.1038/s41592-023-01773-7 (2023).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Zeng, H. et al. Integrative in situ mapping of single-cell transcriptional states and tissue histopathology in a mouse model of Alzheimer’s disease. Nat. Neurosci. 26, 430–446 (2023).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods https://doi.org/10.1038/s41592-021-01358-2 (2022).
Hu, J. et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Fang, R. et al. Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science 377, 56–62 (2022).
Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell https://doi.org/10.1016/j.cell.2023.06.009 (2023).
Li, Z. & Zhou, X. BASS: multi-scale and multi-sample analysis enables accurate cell type clustering and spatial domain detection in spatial transcriptomic studies. Genome Biol. 23, 168 (2022).
Dong, K. & Zhang, S. Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nat. Commun. 13, 1739 (2022).
Zeira, R., Land, M., Strzalkowski, A. & Raphael, B. J. Alignment and integration of spatial transcriptomics data. Nat. Methods 19, 567–575 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Moving towards reproducible machine learning. Nat. Comput. Sci. 1, 629–630 https://doi.org/10.1038/s43588-021-00152-6 (2021).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science https://doi.org/10.1126/science.aar7042 (2018).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. In Situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. Y. & Zhuang, X. W. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Keren, L. et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. https://doi.org/10.1126/sciadv.aax5851 (2019).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0739-1 (2020).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science https://doi.org/10.1126/science.aat5691 (2018).
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 e915 (2018).
Codeluppi, S. et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods 15, 932–935 (2018).
Sun, S., Zhu, J. & Zhou, X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods 17, 193–200 (2020).
Zhu, J., Sun, S. & Zhou, X. SPARK-X: non-parametric modeling enables scalable and robust detection of spatial expression patterns for large spatial transcriptomic studies. Genome Biol. 22, 184 (2021).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00935-2 (2021).
Shang, L. & Zhou, X. Spatially aware dimension reduction for spatial transcriptomics. Nat. Commun. 13, 7203 (2022).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00830-w (2021).
Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01273-7 (2022).
Anderson, A. & Lundeberg, J. sepal: identifying transcript profiles with spatial patterns by diffusion-based modeling. Bioinformatics https://doi.org/10.1093/bioinformatics/btab164 (2021).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Borm, L. E. et al. Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01455-3 (2022).
Ren, H., Walker, B. L., Cang, Z. & Nie, Q. Identifying multicellular spatiotemporal organization of cells with SpaceFlow. Nat. Commun. https://doi.org/10.1038/s41467-022-31739-w (2022).
Chen, X., Sun, Y.-C., Church, G. M., Lee, J. H. & Zador, A. M. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 46, e22–e22 (2018).
Fu, H. et al. Unsupervised spatial embedded deep representation of spatial transcriptomics. Preprint at bioarxiv https://doi.org/10.1101/2021.06.15.448542 (2021).
Long, B., Miller, J. & Consortium, T. S. SpaceTx: a roadmap for benchmarking spatial transcriptomics exploration of the brain. Preprint at https://doi.org/10.48550/arXiv.2301.08436 (2023).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. https://doi.org/10.1038/s41587-019-0392-8 (2020).
Zhao, T. et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell https://doi.org/10.1016/j.cell.2020.10.026 (2020).
Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 1–12 (2019).
Haghverdi, L., Buttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Network, B. I. C. C. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Tu, J.-J., Li, H.-S., Yan, H., Zhang, X.-F. & Boeva, V. EnDecon: cell type deconvolution of spatially resolved transcriptomics data via ensemble learning. Bioinformatics https://doi.org/10.1093/bioinformatics/btac825 (2023).
Liao, J. et al. De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution. Nat. Commun. https://doi.org/10.1038/s41467-022-34271-z (2022).
Cable, D. M. et al. Cell type-specific inference of differential expression in spatial transcriptomics. Nat. Methods 19, 1076–1087 (2022).
Jerby-Arnon, L. & Regev, A. DIALOGUE maps multicellular programs in tissue from single-cell or spatial transcriptomics data. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01288-0 (2022).
Yuan, Z. et al. SOTIP is a versatile method for microenvironment modeling with spatial omics data. Nat. Commun. https://doi.org/10.1038/s41467-022-34867-5 (2022).
Acknowledgements
This work was supported by by Chenguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission, Shanghai Science and Technology Development Funds (23YF1403000), Tencent AI Lab Rhino-Bird Focused Research Program (RBFR2023008), Shanghai Municipal Science and Technology Major Project (no. 2018SHZDZX01), ZJ Lab, Shanghai Center for Brain Science and Brain-Inspired Technology and 111 Project (no. B18015). The Innovation Fund of Institute of Computing and Technology, CAS (E161080, E161030); Beijing Natural Science Foundation Haidian Origination and Innovation Joint Fund (L222007).
Author information
Authors and Affiliations
Contributions
Y.Z. and Z.Y. conceived and designed the study. Z.Y. designed the pipeline and collected the methods and datasets. S.L. and Z.Y. completed the pipeline. Z.Y. and S.L. analyzed the results and generated the figures. J.Y. and Z.W. maintained the database. Z.Y., S.L., Z.F. and Y.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Yuan, Z. et al. Nat. Methods 20, 387–399 (2023): https://doi.org/10.1038/s41592-023-01773-7
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Supplementary Table 2 and Supplementary Protocols.
Supplementary Table 1
Summary of parameters.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, S., Zhao, F., Wu, Z. et al. Streamlining spatial omics data analysis with Pysodb. Nat Protoc 19, 831–895 (2024). https://doi.org/10.1038/s41596-023-00925-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00925-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.