Abstract
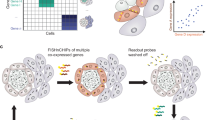
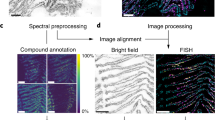
Label-free molecular imaging techniques such as matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI) enable the direct and simultaneous mapping of hundreds of different metabolites in thin sections of biological tissues. However, in host–microbe interactions it remains challenging to localize microbes and to assign metabolites to the host versus members of the microbiome. We therefore developed a correlative imaging approach combining MALDI-MSI with fluorescence in situ hybridization (FISH) on the same section to identify and localize microbial cells. Here, we detail metaFISH as a robust and easy method for assigning the spatial distribution of metabolites to microbiome members based on imaging of nucleic acid probes, down to single-cell resolution. We describe the steps required for tissue preparation, on-tissue hybridization, fluorescence microscopy, data integration into a correlative image dataset, matrix application and MSI data acquisition. Using metaFISH, we map hundreds of metabolites and several microbial species to the micrometer scale on a single tissue section. For example, intra- and extracellular bacteria, host cells and their associated metabolites can be localized in animal tissues, revealing their complex metabolic interactions. We explain how we identify low-abundance bacterial infection sites as regions of interest for high-resolution MSI analysis, guiding the user to a trade-off between metabolite signal intensities and fluorescence signals. MetaFISH is suitable for a broad range of users from environmental microbiologists to clinical scientists. The protocol requires ~2 work days.
Key points
-
A procedure for spatial metabolomics of host–microbe interactions, including tissue preparation, matrix application, MSI data acquisition, on-tissue hybridization using nucleic acid probes, fluorescence microscopy and data integration into a correlative image dataset.
-
MALDI-MSI enables single-cell-level mapping of metabolites by revealing their spatial distribution. Alternatively, laser-capture microdissection can be combined with LC–MS, or metaFISH combines spatial metabolomics with FISH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data presented in this paper have been deposited in the METASPACE project protocol (https://metaspace2020.eu/project/metaFISH). Individual datasets are deposited as follows: Fig. 1, MPIMM_193_QE_P_BC_CF (https://metaspace2020.eu/dataset/2019-11-28_11h08m15s); Fig. 5, 20210518_b_child_nix_s1_dhap_maldi2_tof_5um_laser90%_50shots (https://metaspace2020.eu/dataset/2021-07-08_13h54m26s), 20210518_b_child_nix_s1_dhap_laser70%_shots50 (https://metaspace2020.eu/dataset/2021-05-30_18h51m15s), 20210518_b_child_nix_s1_dhap_5um_maldi2_tof_laser50%_100 (https://metaspace2020.eu/dataset/2021-05-30_18h08m20s), 20210518_b_child_nix_s1_dhap_5um_maldi2_tof_laser30%_150 (https://metaspace2020.eu/dataset/2021-05-30_18h08m14s), and MPIMM_299_TTF_M2_Grid (https://metaspace2020.eu/dataset/2023-01-26_10h37m44s); Fig. 6, 20210706_bchild_nix_n25_tm_sl108 (https://metaspace2020.eu/dataset/2021-07-08_15h03m30s); Fig. 7, MTBLS2639; and Fig. 8, MPIMM_054_QE_P_BP_CF (https://metaspace2020.eu/dataset/2017-03-28_16h40m57s) and MPIMM_193_QE_P_BC_CF (https://metaspace2020.eu/dataset/2019-11-28_11h08m15s).
Code availability
Open-source scripts for the implementation of the MSI and microscopy co-registration have been published previously6 and are available on GitHub (R scripts, https://github.com/esogin/miniature-octo-fiesta; MATLAB, https://github.com/BenediktSenorDingDong/MALDI-FISHregistration).
References
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Mund, A., Brunner, A.-D. & Mann, M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol. Cell 82, 2335–2349 (2022).
Bauermeister, A., Mannochio-Russo, H., Costa-Lotufo, L. V., Jarmusch, A. K. & Dorrestein, P. C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 20, 143–160 (2022).
Wishart, D. S. et al. NMR and metabolomics—a roadmap for the future. Metabolites 12, 678 (2022).
Alexandrov, T. Spatial metabolomics and imaging mass spectrometry in the age of artificial intelligence. Annu. Rev. Biomed. Data Sci. 3, 61–87 (2020).
Geier, B. et al. Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nat. Microbiol. 5, 498–510 (2020).
Esquenazi, E., Yang, Y.-L., Watrous, J., Gerwick, W. H. & Dorrestein, P. C. Imaging mass spectrometry of natural products. Nat. Prod. Rep. 26, 1521–1534 (2009).
Vickerman, J. C. Molecular imaging and depth profiling by mass spectrometry—SIMS, MALDI or DESI. Analyst 136, 2199–2217 (2011).
Pozebon, D., Scheffler, G. L., Dressler, V. L. & Nunes, M. A. G. Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples. J. Anal. At. Spectrom. 29, 2204–2228 (2014).
Heijs, B., Potthoff, A., Soltwisch, J. & Dreisewerd, K. MALDI-2 for the enhanced analysis of N-linked glycans by mass spectrometry imaging. Anal. Chem. 92, 13904–13911 (2020).
Stanback, A. E. et al. Regional N-glycan and lipid analysis from tissues using MALDI–mass spectrometry imaging. STAR Protoc. 2, 100304 (2021).
Sun, C. et al. Visualizing the spatial distribution and alteration of metabolites in continuously cropped Salvia miltiorrhiza Bge using MALDI–MSI. J. Pharm. Anal. 12, 719–724 (2022).
Sogin, E. M. et al. Sugars dominate the seagrass rhizosphere. Nat. Ecol. Evol. 6, 866–877 (2022).
Bien, T., Koerfer, K., Schwenzfeier, J., Dreisewerd, K. & Soltwisch, J. Mass spectrometry imaging to explore molecular heterogeneity in cell culture. Proc. Natl Acad. Sci. USA 119, e2114365119 (2022).
Capolupo, L. et al. Sphingolipids control dermal fibroblast heterogeneity. Science 376, eabh1623 (2022).
Bourceau, P., Michellod, D., Geier, B. & Liebeke, M. Spatial metabolomics shows contrasting phosphonolipid distributions in tissues of marine bivalves. PeerJ Anal. Chem. 4, e21 (2022).
Bowman, A. P. et al. Evaluation of lipid coverage and high spatial resolution MALDI-imaging capabilities of oversampling combined with laser post-ionisation. Anal. Bioanal. Chem. 412, 2277–2289 (2020).
Rappez, L. et al. SpaceM reveals metabolic states of single cells. Nat. Methods 18, 799–805 (2021).
Wang, G. et al. Analyzing cell-type-specific dynamics of metabolism in kidney repair. Nat. Metab. 4, 1109–1118 (2022).
Zhu, X., Xu, T., Peng, C. & Wu, S. Advances in MALDI mass spectrometry imaging single cell and tissues. Front. Chem. 9, 782432 (2022).
Niehaus, M., Soltwisch, J., Belov, M. E. & Dreisewerd, K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 16, 925–931 (2019).
Kompauer, M., Heiles, S. & Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 14, 90–96 (2017).
Yang, J. Y. et al. Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 194, 6023–6028 (2012).
Yang, Y.-L., Xu, Y., Straight, P. & Dorrestein, P. C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 5, 885–887 (2009).
Si, T. et al. Characterization of Bacillus subtilis colony biofilms via mass spectrometry and fluorescence imaging. J. Proteome Res. 15, 1955–1962 (2016).
Feucherolles, M. & Frache, G. MALDI mass spectrometry imaging: a potential game-changer in a modern microbiology. Cells 11, 3900 (2022).
Dunham, S. J. B., Ellis, J. F., Li, B. & Sweedler, J. V. Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res. 50, 96–104 (2017).
Gahlmann, A. & Moerner, W. E. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat. Rev. Microbiol. 12, 9–22 (2014).
Hatzenpichler, R., Krukenberg, V., Spietz, R. L. & Jay, Z. J. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Microbiol. 18, 241–256 (2020).
Amann, R. & Fuchs, B. M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6, 339–348 (2008).
Amann, R., Fuchs, B. M. & Behrens, S. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12, 231–236 (2001).
Barrero-Canosa, J., Moraru, C., Zeugner, L., Fuchs, B. M. & Amann, R. Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ. Microbiol. 19, 70–82 (2017).
Greuter, D., Loy, A., Horn, M. & Rattei, T. probeBase—an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 44, D586–D589 (2015).
Smith, B. et al. Community analysis of bacteria colonizing intestinal tissue of neonates with necrotizing enterocolitis. BMC Microbiol. 11, 73 (2011).
Schimak, M. P., Toenshoff, E. R. & Bright, M. Simultaneous 16S and 18S rRNA fluorescence in situ hybridization (FISH) on LR White sections demonstrated in Vestimentifera (Siboglinidae) tubeworms. Acta Histochem. 114, 122–130 (2012).
Neugent, M. L., Gadhvi, J., Palmer, K. L., Zimmern, P. E. & De Nisco, N. J. Detection of tissue-resident bacteria in bladder biopsies by 16S rRNA fluorescence in situ hybridization. J Vis. Exp. 152, e60458 (2019).
Valm, A. M., Mark Welch, J. L. & Borisy, G. G. CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 35, 496–502 (2012).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Kaltenpoth, M., Strupat, K. & Svatoš, A. Linking metabolite production to taxonomic identity in environmental samples by (MA)LDI–FISH. ISME J. 10, 527–531 (2016).
Geier, B. et al. Connecting structure and function from organisms to molecules in small-animal symbioses through chemo-histo-tomography. Proc. Natl Acad. Sci. USA 118, e2023773118 (2021).
Ponnudurai, R. et al. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 11, 463–477 (2017).
Dreisewerd, K., Bien, T. & Soltwisch, J. in Mass Spectrometry Imaging of Small Molecules: Methods and Protocols (ed. Y.-J. Lee) 21–40 (Springer, 2022).
Soltwisch, J. et al. Mass spectrometry imaging with laser-induced postionization. Science 348, 211–215 (2015).
Ansorge, R. et al. Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nat. Microbiol. 4, 2487–2497 (2019).
Fung, C. et al. High-resolution mapping reveals that microniches in the gastric glands control Helicobacter pylori colonization of the stomach. PLoS Biol. 17, e3000231 (2019).
Lackner, G., Peters, E. E., Helfrich, E. J. N. & Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl Acad. Sci. USA 114, E347–E356 (2017).
Yang, H. et al. On-tissue derivatization of lipopolysaccharide for detection of Lipid A using MALDI-MSI. Anal. Chem. 92, 13667–13671 (2020).
Patel, E. et al. MALDI-MS imaging for the study of tissue pharmacodynamics and toxicodynamics. Bioanalysis 7, 91–101 (2015).
Prideaux, B. et al. High-sensitivity MALDI–MRM–MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal. Chem. 83, 2112–2118 (2011).
Daims, H., Brühl, A., Amann, R., Schleifer, K.-H. & Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444 (1999).
Braun, P. et al. In-depth analysis of Bacillus anthracis 16S rRNA genes and transcripts reveals intra- and intergenomic diversity and facilitates anthrax detection. mSystems 7, e01361–01321 (2022).
Rath, C. M. et al. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal. Chem. 84, 9259–9267 (2012).
Piwosz, K., Mukherjee, I., Salcher, M. M., Grujčić, V. & Šimek, K. CARD-FISH in the sequencing era: opening a new universe of protistan ecology. Front. Microbiol. 12, 640066 (2021).
Morales, D. P. et al. Advances and challenges in fluorescence in situ hybridization for visualizing fungal endobacteria. Front. Microbiol. 13, 892227 (2022).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Ticha, P. et al. A novel cryo-embedding method for in-depth analysis of craniofacial mini pig bone specimens. Sci. Rep. 10, 19510 (2020).
Hoffmann, F., Janussen, D., Dröse, W., Arp, G. & Reitner, J. Histological investigation of organisms with hard skeletons: a case study of siliceous sponges. Biotech. Histochem. 78, 191–199 (2003).
Kompauer, M., Heiles, S. & Spengler, B. Autofocusing MALDI mass spectrometry imaging of tissue sections and 3D chemical topography of nonflat surfaces. Nat. Methods 14, 1156–1158 (2017).
Angerer, T. B., Bour, J., Biagi, J.-L., Moskovets, E. & Frache, G. Evaluation of 6 MALDI–matrices for 10 μm lipid imaging and on-tissue MSn with AP-MALDI-Orbitrap. J. Am. Soc. Mass Spectrom. 33, 760–771 (2022).
Leopold, J., Prabutzki, P., Engel, K. M. & Schiller, J. A five-year update on matrix compounds for MALDI–MS analysis of lipids. Biomolecules 13, 546 (2023).
Cerruti, C. D., Benabdellah, F., Laprévote, O., Touboul, D. & Brunelle, A. MALDI imaging and structural analysis of rat brain lipid negative ions with 9-aminoacridine matrix. Anal. Chem. 84, 2164–2171 (2012).
Kaya, I., Jennische, E., Lange, S. & Malmberg, P. Dual polarity MALDI imaging mass spectrometry on the same pixel points reveals spatial lipid localizations at high-spatial resolutions in rat small intestine. Anal. Methods 10, 2428–2435 (2018).
Meisenbichler, C., Doppler, C., Bernhard, D. & Müller, T. Improved matrix coating for positive- and negative-ion-mode MALDI-TOF imaging of lipids in blood vessel tissues. Anal. Bioanal. Chem. 411, 3221–3227 (2019).
Shariatgorji, R. et al. Spatial visualization of comprehensive brain neurotransmitter systems and neuroactive substances by selective in situ chemical derivatization mass spectrometry imaging. Nat. Protoc. 16, 3298–3321 (2021).
Iwama, T. et al. Development of an on-tissue derivatization method for MALDI mass spectrometry imaging of bioactive lipids containing phosphate monoester using Phos-tag. Anal. Chem. 93, 3867–3875 (2021).
Harkin, C. et al. On-tissue chemical derivatization in mass spectrometry imaging. Mass Spectrom. Rev. 41, 662–694 (2022).
Tressler, C. et al. Factorial design to optimize matrix spraying parameters for MALDI mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 32, 2728–2737 (2021).
Ščupáková, K. et al. Cellular resolution in clinical MALDI mass spectrometry imaging: the latest advancements and current challenges. Clin. Chem. Lab. Med. 58, 914–929 (2020).
Schimak, M. P. et al. MiL-FISH: multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl. Environ. Microbiol. 82, 62–70 (2016).
Amann, R. I., Krumholz, L. & Stahl, D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770 (1990).
Folberth, J., Begemann, K., Jöhren, O., Schwaninger, M. & Othman, A. MS(2) and LC libraries for untargeted metabolomics: Enhancing method development and identification confidence. J. Chromatogr. B 1145, 122105 (2020).
Chaleckis, R., Meister, I., Zhang, P. & Wheelock, C. E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 55, 44–50 (2019).
Palmer, A. et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods 14, 57–60 (2017).
Spraggins, J. M. et al. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem. 91, 14552–14560 (2019).
Kim, Y. H. et al. In situ label-free visualization of orally dosed strictinin within mouse kidney by MALDI–MS imaging. J. Agric. Food Chem. 62, 9279–9285 (2014).
Pirman, D. A., Reich, R. F., Kiss, A. A., Heeren, R. M. A. & Yost, R. A. Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal. Chem. 85, 1081–1089 (2013).
Amann, R. I., Ludwig, W. & Schleifer, K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 (1995).
Janda, M. et al. Determination of abundant metabolite matrix adducts illuminates the dark metabolome of MALDI-mass spectrometry imaging datasets. Anal. Chem. 93, 8399–8407 (2021).
Protsyuk, I. et al. 3D molecular cartography using LC–MS facilitated by Optimus and ’ili software. Nat. Protoc. 13, 134–154 (2018).
Garg, N. et al. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22, 705–716.e704 (2017).
Shen, S. et al. Parallel, high-quality proteomic and targeted metabolomic quantification using laser capture microdissected tissues. Anal. Chem. 93, 8711–8718 (2021).
Dilillo, M. et al. Mass spectrometry imaging, laser capture microdissection, and LC–MS/MS of the same tissue section. J. Proteome Res. 16, 2993–3001 (2017).
Ščupáková, K., Dewez, F., Walch, A. K., Heeren, R. M. A. & Balluff, B. Morphometric cell classification for single-cell MALDI–mass spectrometry imaging. Angew. Chem. Int. Ed. 59, 17447–17450 (2020).
Prade, V. M. et al. De novo discovery of metabolic heterogeneity with immunophenotype-guided imaging mass spectrometry. Mol. Metabol. 36, 100953 (2020).
Blanc, L., Lenaerts, A., Dartois, V. & Prideaux, B. Visualization of mycobacterial biomarkers and tuberculosis drugs in infected tissue by MALDI–MS imaging. Anal. Chem. 90, 6275–6282 (2018).
Perry, W. J. et al. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc. Natl Acad. Sci. USA 116, 21980–21982 (2019).
Tobias, F. & Hummon, A. B. Considerations for MALDI-based quantitative mass spectrometry imaging studies. J. Proteome Res. 19, 3620–3630 (2020).
Bakker, B. et al. Preparing ductal epithelial organoids for high-spatial-resolution molecular profiling using mass spectrometry imaging. Nat. Protoc. 17, 962–979 (2022).
Paschke, C. et al. Mirion—a software package for automatic processing of mass spectrometric images. J. Am. Soc. Mass Spectrom. 24, 1296–1306 (2013).
Bemis, K. D. et al. Cardinal: an R package for statistical analysis of mass spectrometry-based imaging experiments. Bioinformatics 31, 2418–2420 (2015).
Race, A. M., Styles, I. B. & Bunch, J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 75, 5111–5112 (2012).
Goracci, L. et al. Lipostar, a comprehensive platform-neutral cheminformatics tool for lipidomics. Anal. Chem. 89, 6257–6264 (2017).
Tortorella, S. et al. LipostarMSI: comprehensive, vendor-neutral software for visualization, data analysis, and automated molecular identification in mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 31, 155–163 (2020).
Treu, A. & Römpp, A. Matrix ions as internal standard for high mass accuracy matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. 35, e9110 (2021).
Sládková, K., Houška, J. & Havel, J. Laser desorption ionization of red phosphorus clusters and their use for mass calibration in time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23, 3114–3118 (2009).
He, J., Mo, D., Chen, J. & Luo, L. Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nat. Protoc. 15, 3361–3379 (2020).
Richardson, D. S. et al. Tissue clearing. Nat. Rev. Methods Primers 1, 84 (2021).
Schramm, T. et al. imzML—a common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteomics 75, 5106–5110 (2012).
Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007).
Bik, L. et al. In vivo dermal delivery of bleomycin with electronic pneumatic injection: drug visualization and quantification with mass spectrometry. Expert Opin. Drug Deliv. 19, 213–219 (2022).
Cuypers, E. et al. ‘On the spot’ digital pathology of breast cancer based on single-cell mass spectrometry imaging. Anal. Chem. 94, 6180–6190 (2022).
Duperron, S. et al. A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ. Microbiol. 8, 1441–1447 (2006).
Zielinski, F. U. et al. Widespread occurrence of an intranuclear bacterial parasite in vent and seep bathymodiolin mussels. Environ. Microbiol. 11, 1150–1167 (2009).
Raskin, L., Stromley, J. M., Rittmann, B. E. & Stahl, D. A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60, 1232–1240 (1994).
Manz, W., Amann, R., Ludwig, W., Wagner, M. & Schleifer, K.-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15, 593–600 (1992).
Hayasaka, T., Goto-Inoue, N., Masaki, N., Ikegami, K. & Setou, M. Application of 2,5-dihydroxyacetophenone with sublimation provides efficient ionization of lipid species by atmospheric pressure matrix-assisted laser desorption/ionization imaging mass spectrometry. Surf. Interface Anal. 46, 1219–1222 (2014).
Bien, T., Hambleton, E. A., Dreisewerd, K. & Soltwisch, J. Molecular insights into symbiosis—mapping sterols in a marine flatworm-algae-system using high spatial resolution MALDI-2–MS imaging with ion mobility separation. Anal. Bioanal. Chem. 413, 2767–2777 (2021).
Ellis, S. R., Soltwisch, J., Paine, M. R. L., Dreisewerd, K. & Heeren, R. M. A. Laser post-ionisation combined with a high resolving power orbitrap mass spectrometer for enhanced MALDI–MS imaging of lipids. Chem. Commun. 53, 7246–7249 (2017).
Zhou, Q. et al. A caged in-source laser-cleavable MALDI matrix with high vacuum stability for extended MALDI–MS imaging. Angew. Chem. Int. Ed. 62, e202217047 (2023).
Lukowski, J. K. et al. Storage conditions of human kidney tissue sections affect spatial lipidomics analysis reproducibility. J. Am. Soc. Mass Spectrom. 31, 2538–2546 (2020).
Stoecker, K., Dorninger, C., Daims, H. & Wagner, M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 76, 922–926 (2010).
Pernthaler, A., Pernthaler, J. & Amann, R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101 (2002).
Teixeira, H., Sousa, A. L. & Azevedo, A. S. in Fluorescence In-Situ Hybridization (FISH) for Microbial Cells: Methods and Concepts (eds N. F. Azevedo & C. Almeida) 35–50 (Springer, 2021).
Hugenholtz, P., Tyson, G. W. & Blackall, L. L. in Gene Probes: Principles and Protocols (eds M. A. de Muro & Ralph Rapley) 29–42 (Humana Press, 2002).
Acknowledgements
We acknowledge M. Sadowski (MPI Bremen) and J. Beckmann (MPI Bremen) for technical assistance with FISH and MSI, and Bruker Daltonics for access to timsTOF fleX instrumentation. P.B., B.G. and M.L. thank the Max Planck Society for funding. J.S. and K.D. are grateful for funding from Deutsche Forschungsgemeinschaft (DFG): DR 416/12-1 and SO976/41, SO976/5-1 and DR416/13-1, and CRC TRR332 (Z1). B.G. thanks the Human Frontier in Science Program for postdoctoral funding via a long-term fellowship (LT0015/2022-L).
Author information
Authors and Affiliations
Contributions
P.B., B.G., T.B. and V.S. recorded MSI data. V.S., J.S., T.B. and K.D. assisted in the interpretation of results and writing the manuscript. T.B., V.S. and P.B. conducted the protocol validation experiments. P.B., B.G. and M.L. conceived and designed the study. P.B., B.G. and M.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.B. is an employee of Bruker Daltonics GmbH & Co. KG (Bremen). All other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Laura Sanchez and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Geier, B. et al. Nat. Microbiol. 5, 498–510 (2020): https://doi.org/10.1038/s41564-019-0664-6
Geier, B. et al. Proc. Natl Acad. Sci. USA 118, e2023773118 (2021): https://doi.org/10.1073/pnas.2023773118
Bien, T. et al. Proc. Natl Acad. Sci. USA 119, e2114365119 (2022): https://doi.org/10.1073/pnas.2114365119
Niehaus, M. et al. Nat. Methods 16, 925–931 (2019): https://doi.org/10.1038/s41592-019-0536-2
Supplementary information
Supplementary Information
Supplementary Methods 1–4, Tables 1–13.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bourceau, P., Geier, B., Suerdieck, V. et al. Visualization of metabolites and microbes at high spatial resolution using MALDI mass spectrometry imaging and in situ fluorescence labeling. Nat Protoc 18, 3050–3079 (2023). https://doi.org/10.1038/s41596-023-00864-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00864-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.