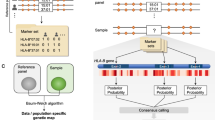
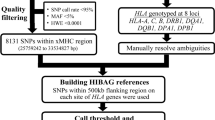
Abstract
The human leukocyte antigen (HLA) locus is associated with more complex diseases than any other locus in the human genome. In many diseases, HLA explains more heritability than all other known loci combined. In silico HLA imputation methods enable rapid and accurate estimation of HLA alleles in the millions of individuals that are already genotyped on microarrays. HLA imputation has been used to define causal variation in autoimmune diseases, such as type I diabetes, and in human immunodeficiency virus infection control. However, there are few guidelines on performing HLA imputation, association testing, and fine mapping. Here, we present a comprehensive tutorial to impute HLA alleles from genotype data. We provide detailed guidance on performing standard quality control measures for input genotyping data and describe options to impute HLA alleles and amino acids either locally or using the web-based Michigan Imputation Server, which hosts a multi-ancestry HLA imputation reference panel. We also offer best practice recommendations to conduct association tests to define the alleles, amino acids, and haplotypes that affect human traits. Along with the pipeline, we provide a step-by-step online guide with scripts and available software (https://github.com/immunogenomics/HLA_analyses_tutorial). This tutorial will be broadly applicable to large-scale genotyping data and will contribute to defining the role of HLA in human diseases across global populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
We have summarized the availability of HLA imputation reference panels in Table 2. Our HLA imputation pipeline using a multi-ancestry HLA reference panel is publicly available at the MIS (https://imputationserver.sph.umich.edu/index.html).
Code availability
The computational scripts and instructions for their usage related to this tutorial are available at https://github.com/immunogenomics/HLA_analyses_tutorial (https://doi.org/10.5281/zenodo.7373439).
References
Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Ann. Rev. Genomics Hum. Genet. 14, 301–323 (2013).
Amiel, J. in Histocompatibility Testing (ed. Teraski, P. I.) 79–81 (Munksgaard, 1967).
Murphy, K. & Weaver, C. Janeway’s immunology. America 1–277 (2017).
Dendrou, C. A., Petersen, J., Rossjohn, J. & Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 18, 325–339 (2018).
Murphy, K. Kenneth M. & Weaver, C. Janeway’s Immunobiology (Garland Science, 2016).
Scally, S. W. et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 210, 2569–2582 (2013).
Ishigaki, K. et al. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet. 54, 393–402 (2022).
McGonagle, D., Aydin, S. Z., Gül, A., Mahr, A. & Direskeneli, H. ‘MHC-I-opathy’-unified concept for spondyloarthritis and Behçet disease. Nat. Rev. Rheumatol. 11, 731–740 (2015).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177 (2016).
Montgomery, R. A., Tatapudi, V. S., Leffell, M. S. & Zachary, A. A. HLA in transplantation. Nat. Rev. Nephrol. 14, 558–570 (2018).
Fleischhauer, K., Zino, E., Bordignon, C. & Benazzi, E. Complete generic and extensive fine-specificity typing of the HLA-B locus by the PCR-SSOP method. Tissue Antigens 46, 281–292 (1995).
Cereb, N., Maye, P., Lee, S., Kong, Y. & Yang, S. Y. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 45, 1–11 (1995).
Erlich, H. HLA DNA typing: past, present, and future. Tissue Antigens 80, 1–11 (2012).
Cereb, N., Kim, H. R., Ryu, J. & Yang, S. Y. Advances in DNA sequencing technologies for high resolution HLA typing. Hum. Immunol. 76, 923–927 (2015).
Smith, A. G. et al. Comparison of sequence-specific oligonucleotide probe vs next generation sequencing for HLA-A, B, C, DRB1, DRB3/B4/B5, DQA1, DQB1, DPA1, and DPB1 typing: toward single-pass high-resolution HLA typing in support of solid organ and hematopoietic cell transplant programs. HLA 94, 296–306 (2019).
Schöfl, G. et al. 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics 18, 1–16 (2017).
Jiao, Y. et al. High-sensitivity HLA typing by saturated tiling capture sequencing (STC-Seq). BMC Genomics 19, 50 (2018).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013).
Dilthey, A. T., Moutsianas, L., Leslie, S. & McVean, G. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27, 968 (2011).
Zheng, X. et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 14, 192–200 (2013).
Luo, Y. et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet. 53, 1504–1516 (2021).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Robinson, J. et al. IPD-IMGT/HLA database. Nucleic Acids Res. 48, D948–D955 (2020).
Marsh, S. G. E. et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291 (2010).
Marsh, S. G. E. et al. An update to HLA nomenclature, 2010. Bone Marrow Transplant. 45, 846–848 (2010).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Dilthey, A. T. et al. High-accuracy HLA type inference from whole-genome sequencing data using population reference graphs. PLoS Comput. Biol. 12, e1005151 (2016).
Dilthey, A. T. et al. HLA*LA—HLA typing from linearly projected graph alignments. Bioinformatics 35, 4394–4396 (2019).
Shen, J. J. et al. HLA-IMPUTER: an easy to use web application for HLA imputation and association analysis using population-specific reference panels. Bioinformatics 35, 1244–1246 (2019).
Maiers, M. et al. GRIMM: GRaph IMputation and matching for HLA genotypes. Bioinformatics 35, 3520–3523 (2019).
Breiman, L. Bagging predictors. Mach. Learn. 24, 123–140 (1996).
Dilthey, A., Cox, C., Iqbal, Z., Nelson, M. R. & McVean, G. Improved genome inference in the MHC using a population reference graph. Nat. Genet. 47, 682–688 (2015).
Hirata, J. et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat. Genet. 51, 470–480 (2019).
Hu, T., Chitnis, N., Monos, D. & Dinh, A. Next-generation sequencing technologies: an overview. Hum. Immunol. 82, 801–811 (2021).
Hosomichi, K., Jinam, T. A., Mitsunaga, S., Nakaoka, H. & Inoue, I. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics 14, 1–16 (2013).
Gibbs, R. A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Browning, S. R. & Browning, B. L. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 12, 703–714 (2011).
Verlouw, J. A. M. et al. A comparison of genotyping arrays. Eur. J. Hum. Genet. 29, 1611 (2021).
Vince, N. et al. SNP-HLA Reference Consortium (SHLARC): HLA and SNP data sharing for promoting MHC-centric analyses in genomics. Genet. Epidemiol. 44, 733–740 (2020).
Klareskog, L., Catrina, A. I. & Paget, S. Rheumatoid arthritis. Lancet 373, 659–672 (2009).
Padyukov, L. et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis. 70, 259–265 (2011).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Wu, P. et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform. https://medinform.jmir.org/2019/4/e14325 (2019).
Gutierrez-Arcelus, M. et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet. 52, 247 (2020).
D’Antonio, M. et al. Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife 8, e48476 (2019).
Aguiar, V. R. C., César, J., Delaneau, O., Dermitzakis, E. T. & Meyer, D. Expression estimation and eQTL mapping for HLA genes with a personalized pipeline. PLoS Genet. 15, e1008091 (2019).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Anderson, C. A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim. 1, 1–21 (2021).
Gilly, A. et al. Very low-depth whole-genome sequencing in complex trait association studies. Bioinformatics 35, 2555–2561 (2019).
Gilly, A. et al. Very low-depth sequencing in a founder population identifies a cardioprotective APOC3 signal missed by genome-wide imputation. Hum. Mol. Genet. 25, 2360–2365 (2016).
Martin, A. R. et al. Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet. 108, 656–668 (2021).
Marees, A. T. et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int. J. Methods Psychiatr. Res 27, e1608 (2018).
Hinrichs, A. S. et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 34, D590–D598 (2006).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Gomes, I. et al. Hardy–Weinberg quality control. Ann. Hum. Genet. 63, 535–538 (1999).
Hosking, L. et al. Detection of genotyping errors by Hardy–Weinberg equilibrium testing. Eur. J. Hum. Genet. 12, 395–399 (2004).
Wittke-Thompson, J. K., Pluzhnikov, A. & Cox, N. J. Rational inferences about departures from Hardy–Weinberg equilibrium. Am. J. Hum. Genet 76, 967 (2005).
Galinsky, K. J. et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 98, 456–472 (2016).
Cook, S. et al. Accurate imputation of human leukocyte antigens with CookHLA. Nat. Commun. 12, 1–11 (2021).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Loh, P.-R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Gourraud, P. A. et al. HLA diversity in the 1000 Genomes Dataset. PLoS One 9, e97282 (2014).
Abi-Rached, L. et al. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS One 13, e0206512 (2018).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Wordsworth, P. et al. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am. J. Hum. Genet. 51, 585 (1992).
Koeleman, B. P. C. et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun. 5, 381–388 (2004).
Thomson, G. et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens 70, 110–127 (2007).
Woelfing, B., Traulsen, A., Milinski, M. & Boehm, T. Does intra-individual major histocompatibility complex diversity keep a golden mean? Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 117–128 (2009).
Lipsitch, M., Bergstrom, C. T. & Antia, R. Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med. Genet. 4, 2 (2003).
Tsai, S. & Santamaria, P. MHC class II polymorphisms, autoreactive T-cells, and autoimmunity. Front. Immunol. 4, 321 (2013).
Goyette, P. et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 47, 172–179 (2015).
Lenz, T. L. et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 47, 1085–1090 (2015).
Arora, J. et al. HLA heterozygote advantage against HIV-1 is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol. Biol. Evol. 37, 639–650 (2020).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015).
Reynolds, E. G. M. et al. Non-additive association analysis using proxy phenotypes identifies novel cattle syndromes. Nat. Genet. 53, 949–954 (2021).
Segal, M. R., Cummings, M. P. & Hubbard, A. E. Relating amino acid sequence to phenotype: analysis of peptide-binding data. Biometrics 57, 632–643 (2001).
Chen, B. et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 37, 1332–1343 (2019).
Pierini, F. & Lenz, T. L. Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol. Biol. Evol. 35, 2145–2158 (2018).
Wakeland, E. K. et al. Ancestral polymorphisms of MHC class II genes: divergent allele advantage. Immunol. Res. 9, 115–122 (1990).
Radwan, J., Babik, W., Kaufman, J., Lenz, T. L. & Winternitz, J. Advances in the evolutionary understanding of MHC polymorphism. Trends Genet. 36, 298–311 (2020).
Chowell, D. et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 25, 1715–1720 (2019).
Choudhury, A. et al. High-depth African genomes inform human migration and health. Nature 586, 741–748 (2020).
Wall, J. D. et al. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576, 106–111 (2019).
Nakane, T. et al. Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pillai, N. E. et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum. Mol. Genet. 23, 4443–4451 (2014).
Okada, Y. et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet. 47, 798–802 (2015).
Zhou, F. et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat. Genet. 48, 740–746 (2016).
Kim, K., Bang, S. Y., Lee, H. S. & Bae, S. C. Construction and application of a Korean reference panel for imputing classical alleles and amino acids of human leukocyte antigen genes. PLoS One 9, e112546 (2014).
Degenhardt, F. et al. Construction and benchmarking of a multi-ethnic reference panel for the imputation of HLA class I and II alleles. Hum. Mol. Genet. 28, 2078–2092 (2019).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021).
Acknowledgements
This work is supported in part by funding from the National Institutes of Health (R01AR063759, U01HG012009, UC2AR081023). S.Sakaue was in part supported by the Manabe Scholarship Grant for Allergic and Rheumatic Diseases, the Uehara Memorial Foundation, and the Osamu Hayaishi Memorial Scholarship. J.B.K. was supported by NIH/NIGMS T32GM007753 and NIH/NIAID F30AI172238. A.J.D. was funded by NIH/NIDDK T32DK007028. T.L.L. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 437857095. Y.O. is supported by AMED (JP22km0405211, JP22km0405217).
Author information
Authors and Affiliations
Contributions
S.Sakaue and S.R. conceived the work and wrote the manuscript with critical input from all authors. S. Sakaue, S.G. and M.C. created a web tutorial accompanying this manuscript. All authors contributed to developing this tutorial. S. Sakaue, M.C., Y.L., W.C., S. Schönherr, L.F., J.L., C.F., Y.O., A.V.S. and S.R. contributed to updating the multi-ancestry HLA reference panel and implementing HLA imputation at the MIS.
Corresponding author
Ethics declarations
Competing interests
B.H. is a CTO of Genealogy Inc. T.L.L. is a co-inventor on a patent application for using HLA evolutionary divergence in predicting cancer immunotherapy success. S.R. is a founder for Mestag, Inc, a scientific advisor for Sonoma, Jannsen and Pfizer, and serves as a consultant for Sanofi and Abbvie.
Peer review
Peer review information
Nature Protocols thanks Judy Cho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The linkage disequilibrium (LD) patterns across the extended MHC region.
A heatmap of LD r2 for pairwise variants across the extended MHC region. We used biallelic markers in our HLA reference panel within European populations and calculated LD r2 values for exhaustive pairs of these variants. The variants are ordered (both on x-axis and y-axis) and annotated by HLA gene names (on x-axis) based on their genomic coordinates on chromosome 6. The bottom plot shows the detailed LD pattern in the class II region.
Extended Data Fig. 2 Schematic illustration of method used to construct scaffold variants within multi-ancestry HLA reference panel.
We extracted SNP variants within MHC region in 1000 Genomes Project (1KG) samples. We only retained variants that were included in major genotyping arrays (Illumina Multi-Ethnic Genotyping Array, Global Screening Array, OmniExpressExome, and Human Core Exome), colored in teal. We then quality controlled each of the participating cohorts’ MHC SNPs separately, retained overlapping variants with selected SNPs in 1KG, and cross-imputed each cohort’s missing variants by using 1KG genotypes. We finally concatenate all cohorts together to construct scaffold variants for multi-ancestry reference panel.
Extended Data Fig. 3 Michigan Imputation Server.
Example usage of Michigan Imputation Server for HLA imputation at https://imputationserver.sph.umich.edu/index.html.
Extended Data Fig. 4 The runtime benchmark for HLA imputation using different platforms.
a. For SNP2HLA, we used BEAGLE4 for phasing and imputation algorithm (Luo et al. Nat Genet. 2021) with using 10 CPUs. For Minmac4, we used SHAPEIT2 as phasing algorithm with samples <10,000 and EAGLE2 as phasing algorithm with samples > 5,000 as we described in the manuscript both with using 10 CPUs. b. For Michigan Imputation Server, we uploaded the unphased genotype data and standard imputation pipeline was performed with default setting (with 1CPU).
Supplementary information
Supplementary Information
Supplementary Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sakaue, S., Gurajala, S., Curtis, M. et al. Tutorial: a statistical genetics guide to identifying HLA alleles driving complex disease. Nat Protoc 18, 2625–2641 (2023). https://doi.org/10.1038/s41596-023-00853-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00853-4
This article is cited by
-
The genetic basis of autoimmunity seen through the lens of T cell functional traits
Nature Communications (2024)
-
Human leukocyte antigen variants associate with BNT162b2 mRNA vaccine response
Communications Medicine (2024)
-
Mapping the dynamic genetic regulatory architecture of HLA genes at single-cell resolution
Nature Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.