Abstract
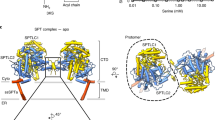
Sphingomyelin (SM) has key roles in modulating mammalian membrane properties and serves as an important pool for bioactive molecules. SM biosynthesis is mediated by the sphingomyelin synthase (SMS) family, comprising SMS1, SMS2 and SMS-related (SMSr) members. Although SMS1 and SMS2 exhibit SMS activity, SMSr possesses ceramide phosphoethanolamine synthase activity. Here we determined the cryo-electron microscopic structures of human SMSr in complexes with ceramide, diacylglycerol/phosphoethanolamine and ceramide/phosphoethanolamine (CPE). The structures revealed a hexameric arrangement with a reaction chamber located between the transmembrane helices. Within this structure, a catalytic pentad E–H/D–H–D was identified, situated at the interface between the lipophilic and hydrophilic segments of the reaction chamber. Additionally, the study unveiled the two-step synthesis process catalyzed by SMSr, involving PE–PLC (phosphatidylethanolamine–phospholipase C) hydrolysis and the subsequent transfer of the phosphoethanolamine moiety to ceramide. This research provides insights into the catalytic mechanism of SMSr and expands our understanding of sphingolipid metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The coordinates are deposited in the Protein Data Bank (PDB) with accession codes 8W9Y, 8IJQ, 8IJR and 8W9W. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-37385, EMD-35492, EMD-35493 and EMD-37383. Other relevant data generated in this study are provided in the Supplementary Information and Source Data files. Source data are provided with this paper.
References
Spector, A. A. & Yorek, M. A. Membrane lipid composition and cellular function. J. Lipid Res. 26, 1015–1035 (1985).
Hannun, Y. A. & Obeid, L. M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018).
Ullman, M. D. & Radin, N. S. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J. Biol. Chem. 249, 1506–1512 (1974).
Slotte, J. P. Biological functions of sphingomyelins. Prog. Lipid Res. 52, 424–437 (2013).
Simons, K. & Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 (2000).
Holthuis, J. C., Pomorski, T., Raggers, R. J., Sprong, H. & Van Meer, G. The organizing potential of sphingolipids in intracellular membrane transport. Physiol. Rev. 81, 1689–1723 (2001).
Huitema, K., van den Dikkenberg, J., Brouwers, J. F. & Holthuis, J. C. Identification of a family of animal sphingomyelin synthases. EMBO J. 23, 33–44 (2004).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017).
Dykstra, M., Cherukuri, A., Sohn, H. W., Tzeng, S. J. & Pierce, S. K. Location is everything: lipid rafts and immune cell signaling. Annu. Rev. Immunol. 21, 457–481 (2003).
Varshney, P., Yadav, V. & Saini, N. Lipid rafts in immune signalling: current progress and future perspective. Immunology 149, 13–24 (2016).
Hannun, Y. A. & Obeid, L. M. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 (2008).
Taniguchi, M. & Okazaki, T. Role of ceramide/sphingomyelin (SM) balance regulated through ‘SM cycle’ in cancer. Cell Signal. 87, 110119 (2021).
Taniguchi, M. & Okazaki, T. Ceramide/sphingomyelin rheostat regulated by sphingomyelin synthases and chronic diseases in murine models. J. Lipid Atheroscler. 9, 380–405 (2020).
Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 18, 33–50 (2018).
Chen, Y. & Cao, Y. The sphingomyelin synthase family: proteins, diseases, and inhibitors. Biol. Chem. 398, 1319–1325 (2017).
Barenholz, Y. & Thompson, T. E. Sphingomyelins in bilayers and biological membranes. Biochim. Biophys. Acta 604, 129–158 (1980).
Yamaoka, S., Miyaji, M., Kitano, T., Umehara, H. & Okazaki, T. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279, 18688–18693 (2004).
Vacaru, A. M. et al. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J. Cell Biol. 185, 1013–1027 (2009).
Ternes, P., Brouwers, J. F., van den Dikkenberg, J. & Holthuis, J. C. Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J. Lipid Res. 50, 2270–2277 (2009).
Yano, M. et al. Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice. J. Biol. Chem. 286, 3992–4002 (2011).
Yano, M. et al. Increased oxidative stress impairs adipose tissue function in sphingomyelin synthase 1 null mice. PLoS One 8, e61380 (2013).
Mitsutake, S. et al. Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, fatty liver, and type 2 diabetes. J. Biol. Chem. 286, 28544–28555 (2011).
Li, Z. et al. Reducing plasma membrane sphingomyelin increases insulin sensitivity. Mol. Cell. Biol. 31, 4205–4218 (2011).
Liu, J. et al. Macrophage sphingomyelin synthase 2 deficiency decreases atherosclerosis in mice. Circ. Res. 105, 295–303 (2009).
Fan, Y. et al. Selective reduction in the sphingomyelin content of atherogenic lipoproteins inhibits their retention in murine aortas and the subsequent development of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30, 2114–2120 (2010).
Qi, X. Y. et al. Discovery of the selective sphingomyelin synthase 2 inhibitors with the novel structure of oxazolopyridine. Bioorg. Med. Chem. Lett. 27, 3511–3515 (2017).
Ding, T. et al. All members in the sphingomyelin synthase gene family have ceramide phosphoethanolamine synthase activity. J. Lipid Res. 56, 537–545 (2015).
Bickert, A. et al. Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice. J. Lipid Res. 56, 821–835 (2015).
Chiang, Y. P., Li, Z., Chen, Y., Cao, Y. & Jiang, X. C. Sphingomyelin synthase related protein is a mammalian phosphatidylethanolamine phospholipase C. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1866, 159017 (2021).
Tafesse, F. G., Ternes, P. & Holthuis, J. C. The multigenic sphingomyelin synthase family. J. Biol. Chem. 281, 29421–29425 (2006).
Cabukusta, B. et al. ER residency of the ceramide phosphoethanolamine synthase SMSr relies on homotypic oligomerization mediated by its SAM domain. Sci. Rep. 7, 41290 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Cabukusta, B., Kohlen, J. A., Richter, C. P., You, C. & Holthuis, J. C. M. Monitoring changes in the oligomeric state of a candidate endoplasmic reticulum (ER) ceramide sensor by single-molecule photobleaching. J. Biol. Chem. 291, 24735–24746 (2016).
Hitosugi, T. et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat. Commun. 4, 1790 (2013).
Hayashi, Y. et al. Carboxyl-terminal tail-mediated homodimerizations of sphingomyelin synthases are responsible for efficient export from the endoplasmic reticulum. J. Biol. Chem. 292, 1122–1141 (2017).
Hayashi, Y. et al. Complex formation of sphingomyelin synthase 1 with glucosylceramide synthase increases sphingomyelin and decreases glucosylceramide levels. J. Biol. Chem. 293, 17505–17522 (2018).
Murakami, C. & Sakane, F. Sphingomyelin synthase-related protein generates diacylglycerol via the hydrolysis of glycerophospholipids in the absence of ceramide. J. Biol. Chem. 296, 100454 (2021).
Chiang, Y. P., Li, Z., Chen, Y., Cao, Y. & Jiang, X. C. Sphingomyelin synthases 1 and 2 exhibit phosphatidylcholine phospholipase C activity. J. Biol. Chem. 297, 101398 (2021).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Yeang, C. et al. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta 1781, 610–617 (2008).
Lou, B. et al. Pharmacologic inhibition of sphingomyelin synthase (SMS) activity reduces apolipoprotein-B secretion from hepatocytes and attenuates endotoxin-mediated macrophage inflammation. PLoS One 9, e102641 (2014).
Adachi, R. et al. Discovery and characterization of selective human sphingomyelin synthase 2 inhibitors. Eur. J. Med. Chem. 136, 283–293 (2017).
Scheres, S. H. W. in The Resolution Revolution: Recent Advances in cryoEM (ed. Crowther, R. A.) Vol 579, 125–157 (Elsevier, 2016).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
We thank M. Lei, D. Ye and L. Wang for scientific discussion and R. Liao for assistance with the data analysis. This work was supported by National Natural Science Foundation of China (grants 82072468 and 82272519 to Y.C.), National Key Research and Development Program of China (grant 2018YFC1004704 to Y.C.), Shanghai Municipal Committee of Science and Technology (grants. 20S11902000 to Y.C. and 21TQ016 to L.Z.), SHIPM-pi fund (grant JY201804 to Y.C. and J.Z.) from Shanghai Institute of Precision Medicine, the Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine. This work was also supported by the Shanghai Frontiers Science Center of Degeneration and Regeneration in Skeletal System (A.Q. and J.Z.) and the Innovative Research Team of High-level Local Universities (grant SHSMU-ZLCX20211700 to Y.C.) from the Shanghai Municipal Education Commission. We thank the staff members of the Electron Microimaging Center, Bioimaging Facility and Proteomics Platform at Shanghai Institute of Precision Medicine for providing technical support and assistance in data collection.
Author information
Authors and Affiliations
Contributions
Y.C. and L.Z. conceived the study. Y.C., L.Z., K.H., Q.Z., B.R., Y.Ch, D.Y., A.Q., J.Z. and S.L. designed the experiments. K.H., Q.Z., Y.X., H.L. and J.Y. performed the biochemical assay. Q.Z., K.H., M.C., Y.S. and Y.C. performed structural biology experiments. Y.C. and D.Y. built and refined structural models. Y.C., K.H., Q.Z., D.Y., J.Y., L.Z., J.Z. and X.J. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Yongchan Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Sara Osman, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed TLC images
Source Data Fig. 2
Unprocessed western blot images
Source Data Fig. 3
Unprocessed TLC images
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Unprocessed TLC images
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Unprocessed TLC images
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Unprocessed TLC images
Source Data Fig. 6
Statistical Source Data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, K., Zhang, Q., Chen, Y. et al. Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01237-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41594-024-01237-2