Abstract
Long interspersed nuclear element 1 (LINE-1) is the only autonomous retrotransposon in humans and new integrations are a major source of genetic variation between individuals. These events can also lead to de novo germline mutations, giving rise to heritable genetic diseases. Recently, a role for DNA repair in regulating these events has been identified. Here we find that Fanconi anemia (FA) DNA crosslink repair factors act in a common pathway to prevent retrotransposition. We purify recombinant SLX4-XPF-ERCC1, the crosslink repair incision complex, and find that it cleaves putative nucleic acid intermediates of retrotransposition. Mice deficient in upstream crosslink repair signaling (FANCA), a downstream component (FANCD2) or the nuclease XPF-ERCC1 show increased LINE-1 retrotransposition in vivo. Organisms limit retrotransposition through transcriptional silencing but this protection is attenuated during early development leaving the zygote vulnerable. We find that during this window of vulnerability, DNA crosslink repair acts as a failsafe to prevent retrotransposition. Together, our results indicate that the FA DNA crosslink repair pathway acts together to protect against mutation by restricting LINE-1 retrotransposition.
Similar content being viewed by others
Main
Long interspersed nuclear element 1 (LINE-1) elements are autonomous transposons in humans and make up 17% of the genome1. LINE-1 retrotransposition occurs by a ‘copy and paste’ mechanism; the RNA intermediate is reverse-transcribed and integrates at a new genomic location2,3,4,5. LINE-1 elements have shaped the evolution of the human genome but are frequently deleterious causing insertional mutagenesis, transcriptional dysregulation and genome instability6,7. New insertions are frequently pathogenic causing inherited diseases or contributing to somatic diseases7,8,9,10,11.
Organisms restrict LINE-1 retrotransposition by limiting its transcription and translation12,13. DNA methylation and chromatin modification play critical roles in restricting LINE-1 (refs. 12,13,14,15). Germline-specific restriction factors, for example the Piwi-interacting RNA (piRNA) system, are critical to restrict LINE-1 and maintain fertility12,14,16. However, recent evidence has implicated DNA repair in promoting and suppressing LINE-1 integration. This suggests that if a LINE-1 element evades transcriptional silencing, DNA repair can limit integration. Factors involved in a wide range of repair processes, but particularly in replication fork stability, have been implicated17,18,19,20,21,22,23. Of interest is the Fanconi anemia (FA) pathway, which repairs DNA interstrand crosslinks (ICLs), as multiple factors from this pathway suppress retrotransposition; however, it is unclear how FA proteins act to suppress LINE-1 retrotransposition13,20,21,24.
While these studies clearly demonstrate that DNA repair can restrict LINE-1 elements in tissue culture, the physiological importance of these restraint mechanisms is unknown. Despite effective retrotransposon transcriptional silencing, new insertions occur both in somatic cells and in the germline; therefore, DNA repair could act to limit retrotransposition in vivo16. Physiological processes required for development render cells more susceptible to LINE-1 integration9,16,24. Perhaps the best examples of this are early zygote and germ cell development in which genome-wide epigenetic reprogramming occurs and LINE-1 transcriptional silencing is attenuated25,26,27. Therefore, DNA repair may play a particularly important role in these situations.
We use reverse genetics to show that DNA crosslink repair factors act in a common pathway to restrict LINE-1 retrotransposition. Moreover, we find that a reconstituted recombinant FA repair incision complex can cleave putative intermediates of retrotransposition. We show that all stages of DNA crosslink repair, defective in the human disease FA, are required to prevent retrotransposition in mice. DNA crosslink repair-deficient mice accumulate LINE-1 integrations in an array of somatic tissues with male germ cells having the highest levels. Finally, we find that early zygotic development is particularly dependent on DNA crosslink repair to prevent LINE-1 retrotransposition.
Results
DNA repair promotes or restrains LINE-1 retrotransposition
Previous studies have identified DNA repair factors that either promote or suppress LINE-1 retrotransposition13,19,28,29. It is proposed that DNA double-strand break (DSB) repair and replication fork stability factors play critical roles in suppressing retrotransposition21.
We confirm these previous results but also ask if other classes of DNA repair factors regulate LINE-1 retrotransposition. We used a previously reported LINE-1 retrotransposition assay in the K562 cell line used to perform LINE-1 genome-wide screens4,13. It is an advantage that all mutants were generated from one parental line, allowing comparison between different mutants. We used genetic knockouts rather than small-interfering RNA allowing quantitative comparisons between mutants but also the generation of double mutants to perform classical genetic studies. A doxycycline (DOX)-inducible promoter drives the expression of the LINE-1 cassette; hence, cells will only be G418 resistant after retrotransposition (Fig. 1a). We showed that LINE-1 retrotransposition was dependent on DOX treatment (Fig. 1b,c). We then used CRISPR–Cas9 to generate cell lines deficient in factors required for DNA DSB repair (Supplementary Fig. 1–4). We first disrupted DNA-PKcs and LIGASE IV, components of nonhomologous end joining (NHEJ) and found that the LINE-1 integration frequency was reduced in agreement with previous reports (Fig. 1d and Supplementary Fig. 1a–f)30,31,32.
a, Schema of the LINE-1 reporter (L1-G418R) used in K562 cells and the strategy used to quantify retrotransposition events. Cells carry a LINE-1 reporter with a G418 resistance cassette in the opposite orientation to the LINE-1 element. The G418 cassette is interrupted with an intron in the orientation of the LINE-1 element. Therefore, the LINE-1 transcript will undergo splicing and, following integration, the noninterrupted G418 cassette will be expressed. b, Image of K562 L1-G418R colonies grown in semisold methylcellulose media following treatment with DOX and selection with G418. c, Quantification of K562 L1-G418R G418-resistant colonies following DOX treatment. n = 6 independent experiments. d–i, As in c but for indicated mutants. Each dot corresponds to an independent experiment. d, NHEJ mutants: ΔDNA-PKcs (sg1), ΔDNA-PKcs (sg2), ΔLIGASEIV (sg1), ΔLIGASEIV (sg2). e, Homologous recombination (HR) mutants: ΔRAD51C, ΔXRCC2, ΔXRCC3, ΔRAD54L, ΔBRCA1, ΔBRCA2. f, BLM: ΔBLM (sg1), ΔBLM (sg2). g, Translesion synthesis (TLS): ΔREV1, ΔREV7. h, Nucleases: ΔSNM1A, ΔSNM1B, ΔFAN1, ΔFEN1, ΔSLX1, ΔMUS81, ΔXPF. i, FA DNA interstrand crosslink repair (FA ICL repair): ΔFANCD2, ΔSLX4. DNA repair mutant K562 cells were generated by CRISPR–Cas9 gene disruption. Sg1 and sg2 represent sgRNAs targeting different exons of the gene of interest. Each dot represents an independent experiment. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by a two-tailed Mann–Whitney U-test (NS, no significant P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
We disrupted RAD51C, XRCC2 and XRCC3, three RAD51 paralogues critical for homologous recombination (Supplementary Fig. 2a,b,e,g–j). LINE-1 integration frequency was increased in the absence of RAD51C but was unaffected by the loss of either XRCC2 or XRCC3 (Fig. 1e). Similar to RAD51C, the loss of BRCA1, BRCA2 or RAD54L led to an increase in the frequency of integration events (Fig. 1e and Supplementary Fig. 2c,d,f). It has been proposed that BRCA1 restricts LINE-1 retrotransposition by protecting the stability of the replication fork17,21. We have expanded on these findings to show that this requirement is not generalizable to all homologous recombination components. Moreover, we tested whether Bloom syndrome helicase (BLM), which can resolve the Holliday junction intermediate of homologous recombination, prevented retrotransposition. We found that the loss of BLM led to a decrease in LINE-1 integration frequency consistent with the hyper-recombinogenic nature of BLM-deficient cells (Fig. 1f and Supplementary Fig. 1g–i)33.
We tested whether other replication fork stability factors suppress retrotransposition. A critical mechanism used by cells to prevent replication fork collapse when impediments to replication are encountered is known as DNA damage tolerance34. Hence, we generated K562 cells deficient in REV1 and REV7 (Supplementary Fig. 3a–c). Loss of either of these factors led to increased LINE-1 integration frequency (Fig. 1g), suggesting there is a general requirement for factors involved in replication fork protection to prevent LINE-1 retrotransposition.
The endonuclease activity of LINE-1 ORF2p is required for efficient retrotransposition; however, when this activity is lost, retrotransposition occurs with much lower efficiency31,32. We generated DNA repair-deficient K562 cell lines in an ORF2p endonuclease-deficient background (Extended Data Fig. 1 and Supplementary Fig. 5). We found that BRCA1 and BRCA2 suppress ORF2p endonuclease-independent integrations as previously reported (Extended Data Fig. 1c)21. REV1 and REV7 were both required to prevent such events (Extended Data Fig. 1d). This agrees with the proposal that LINE-1 could use either the free 3′ OH group of an Okazaki fragment to initiate reverse transcription, or breaks that occur when replication forks collapse17.
As nucleases convert stalled replication forks into breaks, we asked what effect their loss would have on LINE-1 integration. We generated cell lines lacking nucleases required for the maintenance of genome stability (Supplementary Fig. 3d–n). Four nucleases limited nuclease proficient LINE-1 retrotransposition: SNM1A, FAN1, MUS81 and XPF (Fig. 1h). We generated the same repair mutants in the ORF2p endonuclease-dead background (Supplementary Figs. 3l and 5g–j). To our surprise, rather than being required for these events FAN1, MUS81 and XPF limited ORF2p endonuclease-independent integrations (Extended Data Fig. 1f). This reveals that a subset of DNA repair-associated endonucleases is involved in LINE-1 nucleic acid metabolism. Rather than driving retrotransposition in the absence of LINE-1 endonuclease activity or converting stalled replication intermediates into substrates for retrotransposition, these nucleases actually reduce the frequency of LINE-1 integration events.
XPF-deficient cells exhibited an increase in the LINE-1 integration frequency (Fig. 1h). XPF is the nuclease component of the XPF-ERCC1 heterodimeric structure-specific endonuclease critical for multiple DNA repair transactions35. Biochemically, XPF-ERCC1 exhibits nuclease activity on splayed arms, 3′ overhangs and replication fork-like structures36. It was therefore surprising that other nucleases with similar biochemical activities (for example, SLX1) did not limit retrotransposition (Fig. 1h).
This led us to consider how XPF-ERCC1 was channeled into the suppression of LINE-1. XPF functions in multiple repair pathways; however, we found that FANCD2, SLX4 and RAD51C suppressed LINE-1 integration. It was previously hypothesized that FANCD2 protects the replication fork by limiting access to 3′ OH groups on the lagging strand thereby suppressing LINE-1 integration17. Additionally, SLX4 was shown to prevent the accumulation of LINE-1 intermediates in the cytoplasm and induction of cGAS-STING20. As these four factors act in FA DNA ICL repair, it is plausible that this common function is important to limit LINE-1 retrotransposition.
Interstrand crosslink repair suppresses retrotransposition
XPF-ERCC1 is critical for both nucleotide excision repair (NER) and FA repair37,38,39. We formally tested which role of XPF-ERCC1 restricts LINE-1 retrotransposition. We focused our attention on pathway-specific adapters that segregate the nuclease activity into distinct pathways: XPA for NER and SLX4 (FANCP) for FA crosslink repair40,41. We generated two independent mutants of XPA and SLX4 in addition to a further XPF line (Supplementary Fig. 3l,m and 4c,d,g,h). We found the frequency of LINE-1 integration events in XPA-deficient cells is indistinguishable from wild type but SLX4-deficient cells have increased LINE-1 integrations (Fig. 2a,b). In agreement with our findings, in three different genome-wide screens for LINE-1 regulators, XPA was not detected as a suppressor of retrotransposition13,21,24. However, NER has previously been shown to limit retrotransposition; it is plausible that differences in the cell lines used between these studies may explain this discrepancy19. These data indicate that it is the role of XPF-ERCC1 in crosslink repair that is critical to restricting LINE-1 retrotransposition.
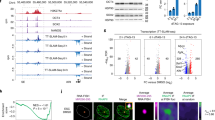
a, Image of K562 L1-G418R wild-type and mutant colonies grown in semisold methylcellulose media following DOX treatment and G418 selection. b, L1 retrotransposition measurement using K562 L1-G418R wild-type and mutant cells (sg1 and sg2, two different sgRNAs). Each dot corresponds to an independent experiment. c, Representative western blot of FANCD2 following 48 h expression of L1-G418R in K562 cells and 24 h MMC treatment, n = 3 independent experiments, P values were calculated by a two-tailed unpaired t-test. d, Representative images of γ-H2A.x foci in K562 L1-G418R cells before and after 48 h DOX treatment. Scale bars, 10 μm. e, Representative images of 53BP1 foci in K562 L1-G418R cells before and after 48 h DOX treatment. Scale bar, 10 μm. f, Percentage of K562 L1-G418R cells with more than five foci. Each dot corresponds to an independent experiment. g, Percentage of K562 L1-G418R cells with more than five foci. Each dot corresponds to an independent experiment. h, Image of K562 L1-G418R wild-type and mutant colonies grown in semisold methylcellulose media following DOX treatment and G418 selection. i, L1 retrotransposition measurement using K562 L1-G418R wild-type and mutant cells (sg1 and sg2, two different sgRNAs), n = 18 independent experiments. j, L1 retrotransposition measurement using L1-G418R in K562 cells (wild-type, FANCD2 mutant and FANCD2 complemented cell lines). Each dot represents data from one independent experiment. k, Schema of the pJJ101/L1.3 reporter used in human fibroblasts. l–n, L1 retrotransposition measurement using pJJ101/L1.3 reporter in: PD220 (FANCA−/−) and complemented line (l), PD331 (FANCC−/−) and complemented line (m) and PD20 (FANCD2−/−) and complemented line (n), n = 3 independent experiments, P values were calculated by a two-tailed unpaired t-test. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by a two-tailed Mann–Whitney U-test.
FA DNA crosslink repair is a highly regulated process with an upstream E3-ubiquitin ligase signaling module that modifies FANCD2. Mono-ubiquitinated FANCD2 promotes incisions at sites of damaged DNA42. We found that the mono-ubiquitination of FANCD2 increased on DOX-induced expression of LINE-1 in K562 cells. This induction was comparable to that induced with the DNA crosslinking agent, mitomycin C (MMC). No such induction was observed in K562 cells treated with DOX that did not harbor the LINE-1 cassette (Fig. 2c).
It has previously been shown that LINE-1 activation causes DNA DSBs, replication stress, checkpoint activation and cell cycle arrest21,24,28. Since the expression of LINE-1 led to the activation of crosslink repair and mono-ubiquitination of FANCD2, we hypothesized that crosslink repair may act to prevent LINE-1-induced DNA DSBs. We generated FANCD2-deficient K562 cells (Supplementary Fig. 4e,f), induced the expression of LINE-1 and quantified DNA damage. We measured TP53-binding protein 1 (53BP1, a surrogate marker of DNA DSBs) and phosphorylation of H2A.X (γ-H2A.X, an alternative marker of DSBs) by immunofluorescence. DOX treatment does not lead to increased DNA damage in wild-type L1-G418R K562 cells (Extended Data Fig. 2a,b), but in the absence of FANCD2, an increased proportion of cells had persistent DNA damage marker foci (Fig. 2d–g). These data indicate that in the absence of crosslink repair factors, the expression of LINE-1 leads to DNA damage signaling and genome instability.
We tested whether upstream components of DNA crosslink repair limit retrotransposition. We assessed the frequency of LINE-1 integration events in cells deficient in FANCA, FANCD2 or XPF. FANCA is a critical factor for the assembly of the FA core complex, the ubiquitin ligase responsible for FANCD2 ubiquitination. We found increased LINE-1 integration frequency for each mutant cell in both LINE-1 endonuclease deficient and proficient backgrounds (Fig. 2h,i, Supplementary Fig. 4a,b,e,f and Extended Data Fig. 1e). The frequency of LINE-1 integration events in FANCA-deficient cells was comparable to those in cells lacking FANCD2 or XPF.
These data indicated that the mono-ubiquitination of FANCD2 is critical to prevent LINE-1 retrotransposition. We complemented FANCD2-deficient cells with either wild type or mutant FANCD2 in which the mono-ubiquitinated lysine is mutated to arginine, K561R (Supplementary Fig. 4i,j)43. In contrast to wild type, the K561R mutant was unable to suppress retrotransposition (Fig. 2j). The K561R data strongly link FANCD2 to FA DNA crosslink repair rather than its roles outside this pathway44,45.
We performed fluctuation analysis to determine the rate of retrotransposition in 12 critical mutants (Extended Data Fig. 3a–g) and confirmed the results presented above (Fig. 1d–i and 2a,b,h,i)46. The loss of repair factors did not alter the expression of the LINE-1 ORF1p and ORF2p proteins (Extended Data Fig. 3h). We used previously described immortalized human fibroblasts from human patients with FA in complementation groups A, C and D2 (PD220, PD331 and PD20, respectively) and their complemented lines to generalize our results beyond K562 cells (Extended Data Fig. 3i–k)47,48. We used a previously reported plasmid-based retrotransposition assay (Fig. 2k)31,49 and found that the loss of FANCA, FANCC or FANCD2 led to increased retrotransposition in patient-derived immortalized primary fibroblasts (Fig. 2l–n). These data provide strong evidence that the entire FA crosslink repair pathway—upstream signaling (core complex components), FANCD2 mono-ubiquitination and downstream incision complex (SLX4 and XPF)—are all required to suppress LINE-1 retrotransposition.
A common pathway for ICL factors to limit retrotransposition
We hypothesized that FA crosslink repair factors act in a common pathway to limit LINE-1 retrotransposition (Fig. 3a). To test this, we generated double knockouts and assessed the frequency of retrotransposition (Supplementary Figs. 3l and 4a,e). We tested whether XPF and SLX4, its regulator in ICL repair, genetically interact and found that the frequency of integration events in the double-mutant was indistinguishable from either single mutant. This epistatic interaction suggests that both factors act together to restrict LINE-1 retrotransposition (Fig. 3b). Second, we tested the interaction between XPF and FANCA and found they were epistatic with respect to LINE-1 retrotransposition (Fig. 3c). Finally, we asked whether the downstream component and substrate of the core complex, FANCD2, genetically interacted with XPF and found a similar epistatic interaction (Fig. 3d). Together, these data strongly indicate that all these factors act in a common pathway to restrict LINE-1 activity.
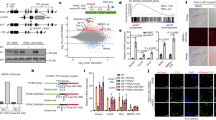
a, Schema illustrating the role of FA DNA ICL repair in restricting L1 retrotransposition. b–d, L1 retrotransposition measurement using L1-G418R in K562 cells testing the genetic interaction between XPF and SLX4 (b), FANCA (c) or FANCD2 (d). e, Schema depicts the SLX4 polypeptide (1–1,834), domains and interactions with nucleases (XPF-ERCC1, MUS81-EME1 and SLX1). A truncated SLX4 1-750 (mini-SLX4) contains the region that interacts with XPF-ERCC1. f, Image of K562 L1-G418R wild-type, SLX4 mutant and SLX4 complemented colonies grown in semisold methylcellulose media following treatment with DOX and selection with G418. g, L1 retrotransposition measurement using L1-G418R in K562 cells (wild-type, SLX4 mutant and complementation cell lines). Each dot represents an independent experiment. h,i, XE and SXE were incubated with different fluorescent-labeled nucleic acid substrates over a time course (collecting samples at 0.2, 5, 10, 20 and 60 min). h, Fork-structured DNA (pseudo-3′ flap). i, Fork-structured DNA with an RNA–DNA hybrid. The reaction products were separated by 12% denaturing PAGE (top), and the decay of the substrate band was quantified and expressed as a percentage of the initial substrate; data were fitted using single-exponential decay (bottom, left) to calculate reaction rates (bottom, right). XE data are plotted in blue; SXE data are plotted in red, n = 2 independent experiments, data represent mean and s.e.m. P values were calculated by a two-tailed Mann–Whitney U-test.
FA incision complex can cleave putative intermediates of retrotransposition
This led us to question how crosslink repair factors suppress LINE-1 retrotransposition. XPF-ERCC1 is a structure-specific endonuclease with the greatest activity on 3′ flaps and replication fork-like substrates36,50. SLX4 enhances the nuclease activity of XPF-ERCC1 toward replication-like structures51. It has been shown that a 3′ overhang is needed to prime the reverse transcription of LINE-1 (ref. 52). Therefore, this first intermediate, proposed in the target-primed reverse transcription (TPRT) model of LINE-1, is likely to be a 3′ flap that is the canonical substrate of SLX4-XPF-ERCC1 (refs. 2,3,4,5).
While we have shown that XPF and SLX4 are epistatic with respect to retrotransposition, SLX4 does interact with two additional nucleases (MUS81 and SLX1)53,54. However, the loss of SLX1 or MUS81 led to negligible increase in retrotransposition (P = 0.0015 and P = 0.0003, respectively, Fig. 1h). Nevertheless, we tested whether the interaction between SLX4 and XPF-ERCC1 was sufficient to suppress retrotransposition. We complemented SLX4-deficient cells with either full-length SLX4 (1–1,834) or mini-SLX4 (1–750) that includes the XPF-ERCC1 binding region (MLR) but does not interact with either SLX1 or MUS81 (Fig. 3e and Extended Data Fig. 4k)40,51,53. Both constructs rescued hypersensitivity to crosslinking agents (Extended Data Fig. 4l) and were able to fully suppress the increase in LINE-1 retrotransposition (Fig. 3f,g). Therefore, the interaction between SLX4 and XPF-ERCC1 is critical to prevent LINE-1 retrotransposition.
Furthermore, we tested whether the nuclease activity of XPF was required to limit retrotransposition. We overexpressed wild type or catalytically dead (D715A) XPF in XPF-deficient cells (Extended Data Fig. 4a–e)55,56. Wild-type XPF suppressed the increase in retrotransposition but the catalytically dead mutant retained high levels of retrotransposition comparable to the XPF-null (Extended Data Fig. 4f). We generated ERCC1-deficient cells and found that they had elevated levels of retrotransposition (Extended Data Figs. 3a and 4g,i,j). We then generated ΔXPFΔERCC1 double mutants and found that they were epistatic with respect to LINE-1 retrotransposition (Extended Data Fig. 4h,j).
We purified recombinant XPF-ERCC1 (XE) and mini-SLX4-XPF-ERCC1 (SXE) (Fig. 3e and Extended Data Fig. 5a) from insect cells to test whether they were able to cleave intermediates of LINE-1 retrotransposition51. As previously reported, we found that the activity of XE on a 3′ pseudoflap was enhanced by SLX4 (Fig. 3h). We next assessed the activity on a 3′ DNA flap with RNA annealed, designed to mimic the intermediate formed during TPRT. As a control, we also annealed DNA generating a double-stranded DNA 3′ branching structure (Extended Data Fig. 5b,c). In both cases, we found that XE cleavage was enhanced by SLX4 (Fig. 3i and Extended Data Fig. 5d–i). We also found that while both XE and SXE nicked double-stranded DNA, neither was able to nick the RNA–DNA hybrid (Extended Data Fig. 5d–i). As the length of the gap between the 3′ end of the RNA and the junction of the splayed DNA arms increased, the activity of XE and SXE was enhanced (Extended Data Fig. 5g–i). This may be due to steric hindrance akin to the inhibition of XPF-ERCC1 activity when a 3′ DNA end invades50,57. We have tested this on a small subset of the potential intermediates of LINE-1 retrotransposition, and SLX4-XPF-ERCC1 may show activity toward others. However, these data indicate a possible mechanism by which these factors repress LINE-1 retrotransposition. The SXE complex could cleave the 3′ DNA flap intermediate aborting integration. Akin to crosslink repair, we propose that the FA ICL repair machinery is recruited to the site of LINE-1 integration, orchestrating and promoting the incision of an intermediate of retrotransposition, aborting new integration events (Fig. 3a).
ICL repair factors inhibit LINE-1 retrotransposition in vivo
FA crosslink repair factors can coordinate cleavage of LINE-1 intermediates and limit retrotransposition in cells but it is unknown whether this is physiologically important. Tracking LINE-1 integrations in vivo is difficult due to their abundance (approximately 600,000 copies), the extremely low rate of retrotransposition and the occurrence of new integrations at different sites in different cells. Newkirk et al. developed a new LINE-1 reporter, the SN1 LINE-1 reporter, that uses the native 5′ untranslated region to drive the expression of the codon-optimized ORF1p and ORF2p, and an enhanced green fluorescent protein (eGFP) cassette in the opposite orientation interrupted by an intron to allow detection and quantification of retrotransposition events (Fig. 4a)58,59. This reporter has limitations, for example the codon-optimization improves expression and increases translation. However, this reporter has indistinguishable DNA methylation and RNA expression dynamics from endogenous LINE-1 elements. Furthermore, the piRNA system that regulates LINE-1 activity also regulates the activity of this reporter59. For these reasons, we chose to use this system for our in vivo assays.
a, SN1 LINE-1 reporter before and after retrotransposition. ORF, open reading frame. UTR, untranslated region. b, Representative ddPCR plots showing frequency of spliced eGFP and Hprt control from skin of wild-type and mutant mice. c, Frequency of spliced eGFP in a skin sample of wild-type and mutant mice by ddPCR. Each point represents one mouse. d, Frequency of spliced eGFP in a skin sample from wild-type and Fancd2−/K559R adult mice by ddPCR. Each point represents one mouse. e, Frequency of spliced eGFP in various tissues from wild-type and Fanca−/− adult mice by ddPCR. Each point represents one mouse. f, Representative flow cytometry plot of kidney single-cell suspension of SN1 and SN1 Fanca−/− mice. g, GFP+ cells in kidney of SN1 and SN1 Fanca−/− mice by flow cytometry. Each point represents one mouse. h, Representative flow cytometry plot of testis single-cell suspension derived from the SN1 and SN1 Fanca−/− mice. i, GFP+ cells in the testis of SN1 and SN1 Fanca−/− mice by flow cytometry. Each point represents one mouse. j, Frequency of spliced eGFP in epididymal sperm from wild-type and Fanca−/− mice by ddPCR. Each point represents one mouse. k, New SN1 LINE-1 integrations in male gametes can be potentially passed onto the next generation. Data represent mean and s.e.m. P values were calculated by a two-tailed Mann–Whitney U-test.
To test the role of ICL repair factors in vivo, we used previously characterized Fanca-, Fancd2- or Ercc1-deficient mice60,61,62. Fanca- and Fancd2-deficient mice are models of the human disease FA with reduced frequencies of blood stem cells, increased cancer predisposition and infertility60,63. Ercc1-deficient mice have a more severe, progeroid phenotype, succumbing to liver failure64,65. All three mutants are infertile due to failure of the embryonic development of primordial germ cells62. We crossed these repair mutants with the SN1 LINE-1 reporter and quantified the frequency of spliced eGFP using a highly sensitive droplet digital (dd)PCR assay (Fig. 4a). We found that we could detect spliced eGFP copies in skin biopsies of DNA repair-proficient mice, but that the frequency was significantly increased in Fanca−/−, Fancd2−/− or Ercc1−/− adults (7.3-, 6.1- and 11.1-fold, respectively, Fig. 4b,c). We tested the requirement for the FANCD2 ubiquitination site by generating a mouse line carrying a K559R mutation (equivalent to K561R in human). Consistent with our in vitro data, we found that Fancd2−/K559R mice had an elevated frequency of spliced eGFP, comparable to Fancd2−/− mice (Fig. 4c,d).
The activation of LINE-1 can induce cGAS-STING signaling66. We hypothesized that cGAS-STING signaling may contribute to the phenotype of Ercc1−/− mice. LINE-1 expression (L1-G418R) in the human K562 cell line activated cGAS-STING and interferon (Extended Data Fig. 6a). Similarly, we observed an induction of cGAS-STING and interferon responses in Ercc1−/− mouse tissues (Extended Data Fig. 6b). We then generated Ercc1−/−Cgas−/− mice to test if cGAS drove aspects of the Ercc1−/− phenotype. However, we found that there was no discernible difference between Ercc1−/−Cgas−/− and Ercc1−/− mice. They had comparable longevity, growth retardation and histological defects (Extended Data Fig. 6c–j). It is therefore unlikely that cGAS-STING signaling plays a major role in the pathogenesis of the Ercc1−/− mice phenotype.
We isolated DNA from different tissues of adult mice with different embryonic origins and replicative potentials and found that the loss of Fanca resulted in an increased frequency of spliced eGFP (Fig. 4e). We performed the same analysis on tissues from Ercc1−/−, Fancd2−/− and Fancd2−/K559R mice observing a comparable pattern of elevated spliced eGFP to wild-type controls (Extended Data Fig. 7a–d). The ddPCR approach may detect both new SN1 LINE-1 integrations but may also amplify unintegrated SN1 LINE-1 complementary DNA. We therefore asked whether we could detect eGFP protein as, in analogy to similar reporters, it is thought that expression of eGFP will only occur after retrotransposition (Fig. 4a)29,67,68,69,70. A single-cell suspension of the kidney was analyzed by flow cytometry. We found that in the absence of FANCA, there was a significant increase in the frequency of GFP+ cells (Fig. 4f,g). We extended this analysis to the bone marrow and lung (tissues from which we could generate single-cell suspensions) and again found a significant increase in the frequency of GFP+ cells in the absence of either FANCA or FANCD2 (Extended Data Fig. 8a–f). We could not detect GFP+ cells in the absence of the SN1 LINE-1 reporter (Extended Data Fig. 8i). These data show an increase in the frequency of spliced eGFP and cells expressing GFP in the absence of the FA pathway.
We measured the messenger RNA (mRNA) of SN1 LINE-1 reporter and repair factors to determine whether expression differences could explain the increase in retrotransposition (Supplementary Fig. 6a–c). We found that the liver, intestine and skin had particularly high expression of SN1 LINE-1 (Supplementary Fig. 6b). We found similar expression of Fanca, Slx4 and Fancd2 between tissues (Supplementary Fig. 6c). We next demonstrated that the loss of these repair factors did not change the expression of the SN1 LINE-1 reporter (Extended Data Fig. 9e and Supplementary Fig. 6d). Together, these results demonstrate that crosslink repair plays an important role in suppressing retrotransposition in mice as well as in human cell lines.
If retrotransposition occurs throughout life, we predicted that the frequency of integrations should accumulate with time. We purified peripheral nucleated white blood cells from wild-type or Fanca−/− 3-month-old mice and found that in the absence of Fanca, there was a 25-fold induction in the frequency of spliced eGFP (Extended Data Fig. 7d). We subsequently bled the same animals at 6 and 9 months (Extended Data Fig. 7e). We did not see any further increase suggesting that the role of crosslink repair in limiting retrotransposition is independent of age (Extended Data Fig. 7f). To ask whether the transcription of the reporter changes with age, we measured the expression of the reporter in the peripheral white blood cells at 3 and 9 months (Extended Data Fig. 7g). There was no significant difference between these time points. We then compared this to the fetal liver at E18.5 (a major site of hematopoiesis) and found that there was a much higher expression of the reporter (Extended Data Fig. 7h). This suggests that the reporter may be more heavily transcribed during fetal development than in adult life.
The testes of Fanca−/− mice stood out as the tissue with the highest rate of retrotransposition (Fig. 4e,h,i). This was surprising as the loss of crosslink repair vastly reduces the number of germ cells with Fanca−/− males having a 4.1-fold reduction in the frequency of tubules exhibiting spermatogenesis62. Therefore, when comparing the testes of wild-type and Fanca−/− males, we analyzed tissues with different cellular compositions (Extended Data Fig. 7i,j). To circumvent this, we purified epididymal sperm, isolated genomic DNA and assessed spliced eGFP. These data showed a 60.5-fold increase in the frequency of spliced eGFP events in Fanca-deficient sperm when compared to littermates (Fig. 4j). Furthermore, the spliced eGFP frequency in sperm was significantly higher than in skin, suggesting that the male germline may be particularly vulnerable to LINE-1 retrotransposition. This is plausible as germ cells undergo complex epigenetic reprogramming events leading to activation of LINE-1 expression and retrotransposition16. We also observed increased rates of spliced eGFP in Fancd2−/− adult testes and Ercc1−/− fetal testes (Extended Data Fig. 7k,l and 8g,h). This is of particular importance, as LINE-1 elements that expand in the germline will be passed on to the next generation and potentially cause disease (Fig. 4k)1,71. Here, we identify DNA crosslink repair as a further tier of protection limiting retrotransposition in these uniquely vulnerable cells.
Maternal and zygotic FA ICL repair factors suppress LINE-1
As LINE-1 integrations do not accumulate with age and are comparably increased across different somatic tissues, we hypothesized that crosslink repair factors restrict retrotransposition during embryogenesis. We assessed the frequency of spliced eGFP at day 18.5 of embryonic development (E18.5) (Fig. 5a). We found that Ercc1−/−, Fanca−/−, Fancd2−/− and Fancd2−/K559R embryos had an elevated frequency of spliced eGFP in multiple tissues (Fig. 5a and Extended Data Fig. 9a–d). We assessed the mRNA expression of the SN1 LINE-1 reporter at E18.5 and found no difference between wild-type and Ercc1−/− embryos (Extended Data Fig. 9e). However, it was striking that the expression of the SN1 LINE-1 reporter was significantly higher in all embryonic tissues when compared to the same tissues in adults (Extended Data Fig. 9f). This suggests that the SN1 LINE-1 reporter is most transcriptionally active during embryogenesis. Taken together, this shows that DNA crosslink repair acts to limit LINE-1 retrotransposition during embryonic development.
a, Quantification of frequency of spliced eGFP copies at E18.5 in wild type, Fanca−/−, Fancd2−/−, Fancd2−/K559R or Ercc1−/− by ddPCR. Each point represents one embryo. b, The strategy to collect oocytes from either Ercc1+/− or Ercc1f/f Zp3-Cre females. c, Expression of Ercc1 was quantified by single-cell qPCR in oocytes. Expression was normalized to Gapdh and made relative to Ercc1+/−. Each point represents one oocyte. d, Ercc1−/Δ embryos were assessed when born from Ercc1f/fZp3-Cre females and Ercc1+/− males. The frequencies at E18.5 (top) or E3.5 (bottom) were assessed. P values were calculated by a two-sided Fischer’s exact test. Exp., expected; Obs., observed. e, The frequency of Ercc1−/− embryos was assessed at E18.5 when born from Ercc1+/− female and Ercc1+/− male. P values were calculated by a two-sided Fischer’s exact test. f, Representative ddPCR plots showing the frequency of spliced eGFP and Hprt control in E18.5 embryos from Ercc1−/− or Ercc1−/Δ embryos from Ercc1+/− or Ercc1f/fZp3-Cre mothers, respectively. g, Quantification of frequency of spliced eGFP copies at E18.5 in wild-type Ercc1−/− or Ercc1−/Δ tissues from Ercc1+/− or Ercc1f/f Zp3-Cre mothers, respectively. Each point represents one embryo. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by a two-tailed Mann–Whitney U-test (NS, not significant P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
It has previously been shown that epigenetic reprogramming results in transcriptional activation of LINE-1 in the zygote and that retrotransposition events that occur during early development lead to somatic and germline mosaicism14,72,73,74. This led us to ask whether DNA crosslink repair factors could limit integration during this period. At fertilization, the RNAs and proteins present in the zygote are maternally derived. As homozygous crosslink repair-deficient mice are infertile, we had to use a conditional allele of Ercc1 (ref. 62). We crossed the conditional allele of Ercc1 with the Tg(Zp3-cre)93Knw allele in which Cre recombinase is expressed under the control of the zona pellucida 3 promoter75. This would allow us to generate Ercc1-deficient oocytes: in other words, an ERCC1 maternal deletion76 (Fig. 5b). We assessed whether (1) oocytes from Ercc1+/− females carried Ercc1 mRNA and (2) if this Ercc1 mRNA was lost in the maternal deletion (Ercc1f/f;Zp3-Cre). We performed quantitative PCR with reverse transcription (RT–qPCR) on individual oocytes harvested from either Ercc1+/− or Ercc1f/f;Zp3-Cre superovulated females. First, we found that oocytes from Ercc1+/− females had detectable Ercc1 mRNA (Fig. 5c). This suggests that the Ercc1−/− mutants we previously studied (Figs. 4 and 5a) had maternal Ercc1 mRNA and protein during the first zygotic divisions. Second, this mRNA was lost in oocytes from Ercc1f/f;Zp3-Cre females showing that we are able to use this strategy to generate oocytes lacking Ercc1 mRNA (Fig. 5c).
We next asked whether maternal Ercc1 mRNA safeguarded the development of ERCC1-deficient zygotes. We crossed the maternal ERCC1-deletion females (Ercc1f/f;Zp3-Cre) with Ercc1+/− males to generate maternal-zygotic ERCC1 deleted embryos (Ercc1−/Δ, Fig. 5d). We harvested preimplantation embryos at E3.5 and assessed the frequency of Ercc1−/Δ blastocysts. There was no significant difference between the observed and expected frequency at E3.5 (Fig. 5d). We assessed the frequency of Ercc1−/Δ embryos at E18.5. The expected frequency was 50%; however, the observed frequency was 10.5% (Fig. 5d). This contrasts with Ercc1−/− embryos generated from Ercc1+/− intercrosses in which there was no significant difference between the expected and observed ratios (Fig. 5e). This suggests that maternal-zygotic deletion of ERCC1 leads to a significant decrease in survival during postimplantation embryonic development.
However, our primary aim was to ask if maternal Ercc1 mRNA offers protection against LINE-1 retrotransposition. We found that Ercc1−/Δ embryos generated from Ercc1f/f;Zp3-Cre females (that is, oocytes lacking Ercc1 mRNA) harbored a striking increase in frequency of spliced eGFP (Fig. 5f, g). We measured these events at E18.5 (as at very early time points, for example E3.5, we obtained insufficient DNA to perform the analysis) allowing us to directly compare these data with that obtained for Ercc1−/− embryos from Ercc1+/− intercrosses (Fig. 5a). This comparison showed that, while Ercc1−/− born from Ercc1+/− intercrosses had a 12.6-fold induction in spliced eGFP, Ercc1−/Δ embryos generated from Ercc1f/f;Zp3-Cre females had a much larger 75.5-fold induction (Fig. 5f,g and Extended Data Fig. 10a,b). We wanted to confirm that the difference observed was not due to the difference between the Ercc1− and Ercc1Δ alleles. We therefore generated Ercc1Δ/Δ, embryos from an Ercc1+/Δ intercross and found that they were born at the expected ratio (Extended Data Fig. 10c, right panel). We then compared the SN1 LINE-1 spliced eGFP frequency with that observed in Ercc1−/− (from Ercc1+/− intercrosses) and found no difference (Extended Data Fig. 10c, left panel). Finally, we showed that Zp3-Cre recombinase itself did not affect either embryo survival or the frequency of SN1 spliced eGFP (Extended Data Fig. 10d). Therefore, our data reveal an important role for maternal Ercc1 in suppressing LINE-1 retrotransposition.
Early zygotic development is extremely vulnerable as epigenetic reprogramming causes temporary transcriptional activation of LINE-1 elements resulting in new integrations that can cause disease or be passed on to the next generation. We find that both maternal and zygotic Ercc1 play a role in preventing these LINE-1 integration events. Together, these data show that during this particularly vulnerable phase, DNA crosslink repair acts as a failsafe mechanism to prevent LINE-1 retrotransposition.
Discussion
Several mechanisms limit LINE-1 retrotransposition but historically the role of DNA repair has been underappreciated. Landmark studies identified a role of DNA repair in regulating retrotransposition in cells13,17,18,19,20,21,22,23,24,28,29,30,77,78,79. We build on this, showing that FA DNA ICL repair factors act in a common pathway to restrain LINE-1 retrotransposition. Biochemically, these factors can cleave putative nucleic acid intermediates of retrotransposition, preventing the priming of reverse transcription. We show that crosslink repair factors also limit LINE-1 retrotransposition in vivo under physiological conditions. This allowed us to identify a particular dependence on crosslink repair during male germ cell and early zygotic development. Together, this shows that FA crosslink repair acts as a failsafe mechanism to limit LINE-1 retrotransposition when other mechanisms of restraint are attenuated.
We have added to the rapidly expanding list of DNA repair pathways (that is, homologous recombination and translesion synthesis) required to limit retrotransposition. It is striking that the repair processes identified have well-described roles in maintaining DNA replication and limiting the formation of DNA DSBs. These factors may act directly on replication fork intermediates or alternative DNA structures, for example DNA secondary structures, to limit retrotransposition. Further studies will be essential to explain whether these factors are redundant to each other or are each dedicated to limiting retrotransposition in distinct situations.
It is possible that in the absence of these repair factors, the expression of the LINE-1 reporter (either mRNA or protein) is increased, leading to elevated retrotransposition. However, we did not observe altered expression of ORF1p and ORF2p in the absence of FANCA, FANCD2 or XPF (Extended Data Figs. 3h and 9e and Supplementary Fig. 6d). The FA pathway regulates an incision complex (SLX4-XPF-ERCC1) that cleaves DNA at ICLs to allow lesion bypass and replication to reach completion. It was striking to us that an intermediate of the LINE-1 TPRT model is a 3′ flap: the canonical substrate for XPF-ERCC1 (ref. 36). Our observation that the addition of SLX4 can stimulate XPF-ERCC1 cleavage of these intermediates provides a simple mechanism to limit retrotransposition: namely preventing priming of reverse transcription. However, canonical TPRT is not the only model proposed for LINE-1 integration. It has been proposed that ORF2p may generate staggered nicks on opposite stands, generating a DSB with a 3′ overhang which could act to prime reverse transcription. It has also been suggested that the resection of telomeres could generate 3′ overhangs to start retrotransposition. It is therefore possible that the FA pathway could limit retrotransposition by cleavage of one of these DNA structures that are independent of DNA replication32,80. Finally, LINE-1 could exploit DSBs that are generated following DNA damage and, subsequent to resection, could lead to the generation of a 3′ flap. Indeed, it remains plausible that in the absence of DNA crosslink repair, there is an increased frequency of stalled replication forks and that these forks could be the substrate for LINE-1 retrotransposition either directly (for example, through the persistence of 3′ OH groups in lagging strand synthesis) or following endonuclease-mediated cleavage17.
DNA crosslink repair deficiency causes FA in humans, characterized by developmental defects, bone marrow failure and cancer predisposition81. There are known differences in both the biology of LINE-1 elements and embryonic development between mice and humans that are important caveats to generalizing the findings of this study. There are differences in the timing of zygote genome activation and in the piRNA system that are likely to affect LINE-1 regulation. However, it is difficult to address these differences as there are both technical and ethical difficulties that preclude studying LINE-1 retrotransposition in human physiological situations. Despite this, it is plausible that increased LINE-1 retrotransposition could contribute to the FA phenotype. LINE-1 retrotransposition events occur at differing rates in human cancers. It is striking that squamous cell carcinomas have among the highest numbers of retrotransposition events and are very common in human patients with FA81,82,83. It has been shown that squamous cell carcinomas from patients with FA shows higher levels of gene rearrangement at repetitive transposon sequences but not increased levels of retrotransposition84. Nonetheless, it will be interesting to test whether tumors deficient in the FA pathway have high levels of LINE-1 retrotransposition and if this contributes to carcinogenesis. Going forward, a key question will be to ask whether preventing LINE-1 transcription suppresses aspects of the FA phenotype, however, this will be technically difficult to test in vivo. It will be necessary to limit LINE-1 transcription as it could be the increased burden of LINE-1-mediated DNA damage rather than increased numbers of integration events that cause tissue dysfunction.
This study has wider implications as we identify DNA repair factors as a failsafe to prevent retrotransposition during developmental stages in which canonical mechanisms are less efficient. Germ cells and early development are particularly vulnerable to LINE-1 as physiologically necessary epigenetic reprogramming leads to loss of transcriptional silencing and activation of LINE-1 (refs. 73,74). As a result, de novo retrotransposition in the male germline has been shown to produce disease-causing mutant alleles85. We have identified DNA crosslink repair as an additional tier of protection to limit such events. It has also become clear that early development may in fact be the most critical window when retrotransposition occurs72. This timing allows the new insertion to be passed on to the next generation through the germline but could also lead to somatic mosaicism. We find that maternal and zygotic DNA crosslink repair factors both contribute to preventing these events. This shows that DNA repair may play a critical role during this extremely vulnerable developmental stage.
Methods
Cell culture
K562 cells were grown in Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and penicillin/streptomycin and cultured at 37 °C with 5% CO2. Human fibroblasts were grown in Dulbecco′s modified Eagle medium supplemented with 10% FBS (Gibco) and penicillin/streptomycin, and cultured at 37 °C with 5% CO2. The cell lines used in the study were tested to be mycoplasma free.
Plasmids
pB-tetO-L1-G418R/blast, pEAK8-FANCD2-YFP, pEAK8-FANCD2(K561R)-YFP, pJJ101/L1.3, pJJ101/L1.3(D702A) and pCEP/GFP were previously published and characterized13,49. pB-tetO-L1(D205A)-G418R/blast reporter was generated by mutagenesis from pB-tetO-L1-G418R/blast with Q5 Site-Directed Mutagenesis Kit Protocol (E0554; New England Laboratories). SLX4 (1–1,834) and truncation ‘mini-SLX4’ (1–750) cDNA sequences were amplified by PCR from genomic DNA and cloned into pcDNA 3.1/Zeo(+). XPF and XPF(D175A) cDNA were previously published56. XPF and XPF(D175A) were amplified by PCR from original plasmids and cloned into pLenti-CMV-GFP-Hygro (Addgene catalog no. 17446), where GFP was exchanged for the cDNA by Gibson assembly. FANCA and FANCC cDNAs were PCR amplified and cloned into pLenti-CMV-GFP-Hygro (Addgene catalog no. 17446), where GFP was exchanged for the cDNA by Gibson assembly. All single-guide RNAs used to generate the DNA repair genetic knockouts in K562 were cloned into pKLV2-U6gRNA5(BbsI)-PGKpuro2AmCherry-W (Addgene catalog no. 67977) or lentiGuide-Hygro-eGFP (Addgene catalog no. 99375). For constitutive Cas9 expression in K562 L1(D205A)-G418R line, K562 cells were nucleofected with lentiCRISPRv2-hygro (Addgene catalog no. 98291).
CRISPR–Cas9-mediated gene disruptions in K562 cells and cell lines generation
Guide sequences for each gene disruption can be found in Supplementary Table 1. K562 cells were nucleofected with the vector containing guides by using Lonza 2b nucleofector (T-016 program). They were either plated directly into 96-well plates as single clones with Puromycin (2.5 μg ml−1, Gibco) selection or 2 days posttransfection, GFP+ or mCherry+ cells were single-cell sorted into 96-well plates. After 14 days at 37 °C, individual clones were analyzed for expression of the relevant protein by western blotting, by Sanger sequencing of targeted loci CRISPR deletion and/or sensitivity to DNA damage agents. Supplementary Table 1 contains the primers used to amplify the relevant loci by PCR. PCR products were cloned into Zero Blunt TOPO (ThermoFisher) and sequenced using an SP6 oligo. To generate the K562 cell line carrying the L1 endonuclease-dead reporter, K562 cells (ATCC) were nucleofected with pB-tetO-L1(D205A)-G418R and plated as single cells into 96-well plates with blasticidin (25 μg ml−1, Gibco) selection until colonies were visible, and then tested individual clones for integration of the plasmid with PCR in both ends (oligos in Supplementary Table 2). The K562 L1(D205A)-G418R cell line was nucleofected with a plasmid containing Cas9 and plated as single cells in 96-well plates with Hygromycin B Gold (200 μg ml−1, InvivoGen) and after 14 days individual clones were analyzed by western blotting to confirm Cas9 expression. For FANCD2 and SLX4 complementation, K562 cells were nucleofected with the vector containing the cDNAs by using Lonza 2b nucleofector (T-016 program). They were plated into 96-well plates as single clones with selection. After 14 days, individual clones were analyzed for expression of the relevant protein by western blotting and by sensitivity to MMC. For XPF complementation, K562 cells were lentivirally transduced with the vector containing cDNAs and 48 h later put in antibiotic selection. Cells were analyzed by western blotting and by sensitivity to MMC. For FANCA and FANCC complementation, human fibroblasts (PD20 and PD331) were lentivirally transduced with the vector containing cDNAs and 48 h later put in antibiotic selection. Cells were analyzed by western blotting and by sensitivity to MMC.
Cell viability assays
Cell viability assays in K562 cells and in human fibroblasts were performed in 96-well flat-bottom plates by plating 1,000 cells per well and culturing them with increasing concentrations of MMC, bleomycin, olaparib and methyl methanesulfonate, or exposing them at ultraviolet irradiation. After 4 days, MTS cell viability reagent (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega) was added and plates were incubated at 37 °C for 4 h; absorbance at 492 nm was measured.
L1-G418 retrotransposition assay in K562 cells
Cells were DOX-induced (1 μg ml−1, Sigma-Aldrich, D9891) for 10 days, maintaining 500,000 cells per ml and refreshing the DOX. After DOX induction, cells were recovered in RPMI medium for 24 h and put into six-well plates with semisolid methylcellulose containing the G418 selection (1 mg ml−1, Formedium Ltd, G4185). G418-resistant colonies were counted after 14 days.
L1-G418 retrotransposition assay in human fibroblasts
Here, 80,000 cells were plated in 100 mm culture dishes and transfected the following day with 10 μl of FuGENE 6 (Promega) and 4 μg of plasmid DNA in OptiMEM medium (ThermoFisher) following the manufacturer’s instructions. PD20 (FANCD2−/−), PD220 (FANCA−/−) and PD331 (FANCC−/−) cells were transfected with pJJ101/L1.3, pJJ101/L1.3-D702A and a plasmid containing blasticidin resistance. After 24 h, fresh media was added and replenished every other day. Then, 5 days posttransfection, cells were selected with 10 μg ml−1 blasticidin (ThermoFisher) for 10 days, with media change every 3 days. Colonies were fixed and stained with crystal violet and colonies counted. Cells were transfected also with pCEP4/GFP to determine the transfection efficiency.
Fluctuation analysis
K562 L1-G418R wild-type and mutant lines underwent fluctuation analysis. For each line, 18 independent cultures (with cultures to determine plating efficiency) were established by plating single cells in 96-well plates in RPMI media and 1 μg ml−1 of DOX (replenished every 72 h). After 10 days, each culture was transferred to a single well in a six-well plate containing semisolid methylcellulose media and 1 mg ml−1 G418. For plating efficiency cultures, serial dilutions were prepared and plated without G418. After 10 days, colonies were counted. Analysis was carried out using http://shinyflan.its.manchester.ac.uk/ and the statistical framework described previously46.
Immunoblotting
Western blots were performed as described previously86. The following primary antibodies were used: anti-XPF (1:1,000, D3G8C, Cell Signaling); anti-ERCC1 (1:100, sc-17809, Santa Cruz Biotechnology); anti-FANCA (1:1,000, D1L2Z, 14657, Cell Signaling Technology); anti-XPA (1:1,000, D9U5U, Cell Signaling); anti-FEN1 (1:2,000, Abcam ab109132); anti-SNM1A (1:500, Abcam ab14805); anti-SNM1B (1:500, Proteintech 13203-1-AP); anti-SLX1 (1:120, MRC PPU S701B); anti-XRCC2 (1:500, Proteintech 20285-1-AP), anti-XRCC3 (1:600, Proteintech 18494-1-AP); anti-Cas9 (1:1,000, 7A9-3A3 14697, Cell Signaling); anti-FLAG (1:200, M2 clone, F1804, Sigma-Aldrich); anti-LAMIN B1 (1:500, ab16048, Abcam); anti-α-TUBULIN (1:3,000, T6199, Sigma-Aldrich); anti-β-ACTIN (1:3,000, Abcam ab8227) and anti-VINCULIN (1:2,000, Abcam ab129002: Abcam); anti-histone H3 (1:7500, catalog no. ab1791, Abcam); anti-LINE-1 ORF1p (1:1,000, clone 4H1, catalog no. MABC1152, Merck); anti-LINE-1 ORF1p (1:1,000, MT49, a gift from K. Burns). They were used for western immunoblotting, diluted in 5% w/v BSA, 0.1% Tween-20 TBS and incubated at 4 °C overnight with gentle agitation. Secondary antibodies were diluted in 5% w/v BSA, 0.1% Tween-20 TBS and incubated for 1 h at room temperature with gentle agitation. For FANCD2 detection, cells were treated with 500 ng ml−1 MMC overnight and protein samples run on a 3–8% Tris-Acetate gel (ThermoFisher). Anti-FANCD2 polyclonal antisera87 was used.
Assessment of DNA damage markers by immunofluorescence was performed as described previously86. DOX treated (48 h) and untreated K562 cells were seeded on top of poly-l-lysine coverslips and spun down for 5 min. Cells were washed twice for 5 min with PBS supplemented with 500 μM MgCl2 and 0.5 μM CaCl2 (PBS-S) then fixed with 4% paraformaldehyde (43368, Alfa Aesar) for 20 min and washed with PBS-S twice for 5 min. They were then washed three times in PBS, 1% w/v Triton X-100 for 15 min. Slides were blocked in PBS, 1% w/v BSA, 1% w/v Triton X-100 for 30 min at room temperature before being incubated overnight at 4 °C with the following primary antibodies diluted in blocking buffer: antiphospho-Histone H2A.X (Ser139) (1:1,000, JBW301, Millipore); anti-53BP1 (1:1,000, NB100-304, Novus). Slides were washed three times in PBS, 1% w/v Triton X-100 for 5 m and incubated with the following secondary antibodies diluted in blocking buffer for 1 h at room temperature: antimouse Alexa Fluor 488 (1:1,000, ThermoFisher) and antirabbit Alexa Fluor 594 (1:1,000, ThermoFisher). The slides were washed three times in PBS, 1% w/v Triton X-100 for 5 min and stained with 0.5 μg ml−1 DAPI diluted in PBS for 10 min. Slides were washed once in water and mounted with ProLong Gold anti-fade reagent (P36934, Molecular Probes) and coverslips were placed onto slides. Images were captured using a Zeiss LSM 780 confocal microscope and analyzed with ImageJv2.9.0/1.53t and Fiji88. DNA damage foci per nucleus were scored blindly.
Expression and purification of protein complexes
SF-9 insect cells were obtained from Merck (89070101-1VL) and 2 l of insect Sf9 cells were infected at 1–2 × 106 cells per ml with tertiary recombinant baculovirus. They were grown for 68 h and collected. XE and SXE purification steps were carried out in NENT buffer supplemented with 20 mM Tris pH 8.0, 5 mM TCEP, 150-400 mM NaCl, 10% glycerol and protein inhibitors cocktail. Cells were homogenized in NENT buffer supplemented with 40 M imidazole pH 8.0 and 0.1% NP-40 followed by nickel affinity chromatography on NTA agarose (Qiagen). Proteins were eluted with NENT buffer containing 250 mM imidazole pH 8.0. For SXE complex, an maltose-binding protein affinity step (NEB, E8022L) was included and the complex was eluted with 20 mM maltose. The maltose-binding protein-tag was cleaved with tobacco etch virus protease O/N at 4 °C. Complexes were diluted with NENT buffer to reduce salt to 200 mM NaCl and loaded on HP Heparin column (GE Healthcare) and eluted with a salt gradient. Concentrated samples were purified on HiLoad Superose 6 (GE Healthcare) and combined fractions were flash-frozen in liquid N2.
Nuclease assays
All reactions were carried out in nuclease buffer: 10–50 mM Tris (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 1 mM TCEP (Tris(2-carboxyethyl) phosphine–HCl) solution (Pierce, catalog no. 77730) (0.5 M TCEP, pH 7.0), 0–5% glycerol and 0.1 mg ml−1 BSA (NEB) at 22 °C. Oligonucleotides were labeled with FAM on the 5′ terminus as shown in Fig. 3h,i and Extended Data Fig. 5b,c. Oligos sequences are shown in Supplementary Table 3 and Extended Data Fig. 5b,c. Substrates were purified on 15% denaturing PAGE gel, desalted and annealed by slow cooling from 90 °C. Reactions were initiated by the generation of an equimolar mixture (100 nM) of the given substrate and enzyme XE and SXE. 0.2-, 5-, 10-, 20- and 60-min timepoint reactions were collected and quenched with 80% formamide, 200 mM NaCl, 10 mM EDTA and 0.01% bromophenol blue, and analyzed on 12% denaturing PAGE (1× Tris-borate-EDTA, 7 M urea, 12% 19:1 acrylamide/bis-acrylamide). Gels were scanned by Typhoon PhosphoImager (GE Healthcare). Band intensities were determined using Fiji (ImageJ). Relative substrate depletion was plotted against time and fitted by single-exponential decays using GraphPad Prism v.9. The rates of substrate depletion were plotted into a bar chart to underline the rate enhancement.
Mice
All animal experiments undertaken in this study were approved by the Medical Research Council’s Laboratory of Molecular Biology animal welfare and ethical review body and the UK Home Office under the Animal (Scientific Procedures) Act 1986 (license no. PP6752216). Mouse husbandry was performed as described previously62. Mice were maintained under specific pathogen-free conditions in individually ventilated cages (GM500; Techniplast) on Lignocel FS-14 spruce bedding (IPS) with environmental enrichment at 19–23 °C with light from 07:00 to 19:00 and humidity of 45–65%, and were fed Dietex CRM pellets (Special Diet Services) ad libitum. No animals were wild and no field-collected samples were used. Mice were maintained on a C57BL/6J background. Embryos were used at E3.5 or E18.5 as indicated in the text. Samples were collected from animals at 8–12 weeks as specified in the text. Females used in timed mating experiments were aged between 6 and 18 weeks. The investigators were blinded to the genotypes of animals throughout the study and data were acquired by relying purely on identification numbers. Fancatm1a(EUCOMM)Wtsi (MGI ID 4434431), Fancd2tm1Hou (MGI ID 2673422), Ercc1tm1a(KOMP)Wtsi (MGI ID 4362172), TnrTg(L1-EGFP)SN1Fhg (MGI ID 244330), Tg(Zp3-cre)93Knw (MGI 2176187) and Cgastm1d(EUCOMM)Hmgu (MGI ID 2442261) alleles have been described previously59,60,61,62,75,89.
Embryo isolation
Timed matings were performed as described previously62. Pregnant mice were killed by cervical dislocation at E12.5 or E18.5 and embryos collected. Embryos were dissected and tissues collected and stored at −80 °C. Individual fetal gonads were placed into ice-cold PBS and quantification was performed immediately.
Superovulation of females to obtain oocytes
Females were superovulated by treatment with 0.1 ml of PMS, after 48 h 0.1 ml of hCG and 24 h later oocytes were collected. For blastocyst collection, females were superovulated by treatment with 0.1 ml of PMS, after 48 h, 0.1 ml of hCG and mated with males. Mice were checked for the presence of a copulation plug. At E3.5, pregnant females were killed by cervical dislocation and blastocysts collected.
Genomic DNA extraction from mouse tissues
Genomic DNA was isolated from adult mice and E18.5 embryonic tissues with Gentra Puregene Tissue (Qiagen) following the manufacturer’s instructions.
ddPCR
All reactions contained roughly 60 ng of genomic DNA per 20 μl reactions. Probes and oligos were diluted to achieve a final concentration of 250 and 900 nM, respectively. NcoI-HF (New England Biolabs) was incorporated into the reaction and samples were incubated at 37 °C for 15 min (6.4 units of enzyme per reaction). An eight-well Bio-Rad DG8 Droplet Generator cassette was used to generate the droplets, loading 20 μl of sample and 70 μl of droplet oil. Then, 35 μl were transferred to ddPCR 96-well plate (Bio-Rad) and thermal cycling conditions were set up as follows: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 60 °C for 1 min, 98 °C for 10 min. Samples were analyzed in Bio-Rad QX200 Droplet Reader. Results were analyzed using Bio-Rad QuantaSoft software. All experiments were performed using FAM and VIC-labeled probes. Oligos and probes in Supplementary Table 2.
Quantification of GFP positive cells in SN1 mice
Kidney, lung, femur and testes were isolated from adult mice and placed in cold PBS. To obtain single-cell suspensions from kidney, the organ was chopped into small pieces in Petri dishes and pipetted up and down with PBS. The tissue was recovered from the bottom of the tube and treated in 5 ml of Hank’s buffered saline solution (HBSS) containing 25 mg collagenase II at 37 °C for 45 min. After that, the tissue was filtered through a 70 μm cell strainer, spun down and resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS/2.5% v/v FBS). For lung single-cell suspension, procedure was identical but pieces of tissue were treated with HBSS containing 25 mg collagenase II and 10 μg ml−1 DNase for 1 h. For bone marrow, cells were isolated from tibiae and femurs with FACS buffer and strained through 70 μm cell strainers. Testes were placed in a 100 mm culture dish containing 10 ml of PBS. Testicular tubules were separated from tunica albuginea and dissociated with forceps. Tubules were transferred to 15 ml conical tube containing 5 ml of HBSS with 500 μl of 5 mg ml−1 collagenase IV and 25 μl of 10 mg ml−1 DNase solution. This was incubated for 10 min at 37 °C. Supernatant was discarded and tubules were collected and passed to another conical tube containing 5 ml of HBSS with 500 μl of 5 mg ml−1 collagenase IV, 25 μl of 10 mg ml−1 DNase solution and 25 μl of 10 mg ml−1 hyaluronidase. This was incubated for 10 min at 37 °C, shaking the tube every 2 min. Cells were filtered in a 70 μm strainer, spun down and resuspended in FACS buffer. Single-cell suspensions were analyzed by flow cytometry on LSRII (BD Bioscience) with data acquired in FACSDivav6.5 (BD) and analyzed on FlowJo v.10.1r5 (FlowJo LLC) to calculate the frequency of GFP+ cells.
RT–qPCR and gene expression analysis
Total RNA was extracted from either K562 cells or adult mice and embryonic tissues using RNAeasy Kit (Qiagen) and first-strand cDNA was synthesized using Quantitect Reverse Transcription Kit For cDNA Synthesis (Qiagen) following the manufacturer’s instructions. Total RNA was extracted and cDNA was synthesized from mouse oocytes using Single Cell-to-CT RT–qPCR Kit (ThermoFisher). Real-time qPCR analysis for expression of interferons and interferon-inducible genes in K562 cells and in mouse tissues (oligos in Supplementary Table 2) was performed using Brilliant II SYBR Green QPCR Master Mix (catalog no. 600828, Agilent Technologies) in a Viia7 (ThermoFisher) cycler at 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Mean threshold cycles were determined from three technical repeats using the comparative CT method. All expression levels were normalized to human GAPDH or mouse Gapdh (oligos can be found in Supplementary Table 2). Orf2p Sn1 Line-1 expression (oligos in Supplementary Table 2) and DNA repair factors expression in adult and embryonic mice tissues was measured using TaqMan Fast Advance Gene Expression Master Mix (ThermoFisher) in a Viia7 (ThermoFisher) cycler at 95 °C for 20 s and 40 cycles of 95 °C for 1 s and 60 °C for 20 s. Mean threshold cycles were determined from three technical repeats using the comparative CT method. All expression levels were normalized to mouse Gapdh (4352339E, ThermoFisher). Ercc1 expression was assessed with m00468337_m1 and m00468336_g1, Fanca with Mm01243361_g1 and Slx4 with Mm01342461 probes (ThermoFisher).
Histology
Histological analysis was carried out on tissues that had been fixed in buffered formalin for 24 h. The samples were paraffin-embedded and 4 μm sections were cut before staining with hematoxylin and eosin.
Quantification of primordial germ cells in vivo
Quantification was performed as described previously62. Urogenital ridges of E12.5 embryos were isolated and placed into 150 μl of trypsin solution (2.5 μg ml−1 trypsin (Gibco), 25 mM Tris, 120 mM NaCl, 25 mM KCl, 25 mM KH2PO4, 25 mM glucose, 25 mM EDTA, pH 7.6) and incubated for 10 min at 37 °C. Next, 1 μl of Benzonase (Millipore) was added, followed by disaggregation of the sample by pipetting and incubation for for 5 min at 37 °C. Trypsin was inactivated by adding 1 ml of PBS/5% v/v FBS. Following 10 min of centrifugation at 1,000g, the cell pellet was resuspended in 100 μl of Alexa Fluor 647-conjugated anti-human/mouse SSEA-1 antibody (catalog no. MC-480; BioLegend) diluted 1:100 in staining buffer (PBS/2.5% v/v FBS) and incubated at room temperature for 10 min; 300 μl of staining buffer were added to the cell suspension and samples immediately run on an ECLIPSE analyzer (Sony Biotechnology) and data analyzed using FlowJo v.10.1r5 (FlowJo LLC).
Statistics and reproducibility
The number of independent biological samples and technical repeats (n) is indicated in the figure legends. Unless otherwise stated, data are shown as the mean ± s.e.m. The nonparametric Mann–Whitney U-test was used to determine statistical significance unless otherwise indicated in the figure legends. Analysis was performed in GraphPad Prism v.9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Source data are provided with this paper.
References
Huang, C. R., Burns, K. H. & Boeke, J. D. Active transposition in genomes. Annu. Rev. Genet. 46, 651–675 (2012).
Cost, G. J., Feng, Q., Jacquier, A. & Boeke, J. D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910 (2002).
Gilbert, N., Lutz, S., Morrish, T. A. & Moran, J. V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25, 7780–7795 (2005).
Moran, J. V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996).
Luan, D. D., Korman, M. H., Jakubczak, J. L. & Eickbush, T. H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993).
Burns, K. H. Our conflict with transposable elements and its implications for human disease. Annu. Rev. Pathol. 15, 51–70 (2020).
Mita, P. & Boeke, J. D. How retrotransposons shape genome regulation. Curr. Opin. Genet Dev. 37, 90–100 (2016).
Beck, C. R., Garcia-Perez, J. L., Badge, R. M. & Moran, J. V. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 12, 187–215 (2011).
Iskow, R. C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 (2010).
Lee, E. et al. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012).
Shukla, R. et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111 (2013).
Goodier, J. L. Restricting retrotransposons: a review. Mob. DNA 7, 16 (2016).
Liu, N. et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232 (2018).
Kim, S. et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564–579 (2014).
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998).
Yang, F. & Wang, P. J. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin. Cell Dev. Biol. 59, 118–125 (2016).
Flasch, D. A. et al. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177, 837–851.e28 (2019).
Sultana, T. et al. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol. Cell 74, 555–570 e7 (2019).
Servant, G. et al. The nucleotide excision repair pathway limits L1 retrotransposition. Genetics 205, 139–153 (2017).
Bregnard, C. et al. Upregulated LINE-1 activity in the Fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine 8, 184–194 (2016).
Mita, P. et al. BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells. Nat. Struct. Mol. Biol. 27, 179–191 (2020).
Ariumi, Y. et al. DNA repair protein Rad18 restricts LINE-1 mobility. Sci. Rep. 8, 15894 (2018).
Gasior, S. L., Roy-Engel, A. M. & Deininger, P. L. ERCC1/XPF limits L1 retrotransposition. DNA Repair 7, 983–989 (2008).
Ardeljan, D. et al. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat. Struct. Mol. Biol. 27, 168–178 (2020).
Surani, M. A., Hayashi, K. & Hajkova, P. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 (2007).
Smith, Z. D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012).
Fadloun, A. et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 20, 332–338 (2013).
Gasior, S. L., Wakeman, T. P., Xu, B. & Deininger, P. L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 (2006).
Coufal, N. G. et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl Acad. Sci. USA 108, 20382–20387 (2011).
Suzuki, J. et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5, e1000461 (2009).
Morrish, T. A. et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31, 159–165 (2002).
Morrish, T. A. et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446, 208–212 (2007).
Chaganti, R. S., Schonberg, S. & German, J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl Acad. Sci. USA 71, 4508–4512 (1974).
Sale, J. E. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 5, a012708 (2013).
Manandhar, M., Boulware, K. S. & Wood, R. D. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 569, 153–161 (2015).
Ciccia, A., McDonald, N. & West, S. C. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77, 259–287 (2008).
Mulderrig, L. & Garaycoechea, J. I. XPF-ERCC1 protects liver, kidney and blood homeostasis outside the canonical excision repair pathways. PLoS Genet. 16, e1008555 (2020).
Kashiyama, K. et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 92, 807–819 (2013).
Bogliolo, M. et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92, 800–806 (2013).
Crossan, G. P. et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 43, 147–152 (2011).
Orelli, B. et al. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J. Biol. Chem. 285, 3705–3712 (2010).
Knipscheer, P. et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009).
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001).
Reliene, R., Yamamoto, M. L., Rao, P. N. & Schiestl, R. H. Genomic instability in mice is greater in Fanconi anemia caused by deficiency of Fancd2 than Fancg. Cancer Res. 70, 9703–9710 (2010).
Sato, K. et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 31, 3524–3536 (2012).
Krasovec, R. et al. Measuring microbial mutation rates with the fluctuation assay. J. Vis. Exp. https://doi.org/10.3791/60406 (2019).
Jakobs, P. M. et al. Immortalization of four new Fanconi anemia fibroblast cell lines by an improved procedure. Somat. Cell Mol. Genet. 22, 151–157 (1996).
Timmers, C. et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7, 241–248 (2001).
Kopera, H. C., Moldovan, J. B., Morrish, T. A., Garcia-Perez, J. L. & Moran, J. V. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc. Natl Acad. Sci. USA 108, 20345–20350 (2011).
Abdullah, U. B. et al. RPA activates the XPF-ERCC1 endonuclease to initiate processing of DNA interstrand crosslinks. EMBO J. 36, 2047–2060 (2017).
Hodskinson, M. R. et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol. Cell 54, 472–484 (2014).
Monot, C. et al. The specificity and flexibility of l1 reverse transcription priming at imperfect T-tracts. PLoS Genet. 9, e1003499 (2013).
Fekairi, S. et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 (2009).
Svendsen, J. M. et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 (2009).
Enzlin, J. H. & Scharer, O. D. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053 (2002).
Sabatella, M. et al. Repair protein persistence at DNA lesions characterizes XPF defect with Cockayne syndrome features. Nucleic Acids Res. 46, 9563–9577 (2018).
Rodriguez, K., Wang, Z., Friedberg, E. C. & Tomkinson, A. E. Identification of functional domains within the RAD1.RAD10 repair and recombination endonuclease of Saccharomyces cerevisiae. J. Biol. Chem. 271, 20551–20558 (1996).
Wang, P. J. Tracking LINE1 retrotransposition in the germline. Proc. Natl Acad. Sci. USA 114, 7194–7196 (2017).
Newkirk, S. J. et al. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc. Natl Acad. Sci. USA 114, E5635–E5644 (2017).
Houghtaling, S. et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17, 2021–2035 (2003).
Garaycoechea, J. I. et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489, 571–575 (2012).
Hill, R. J. & Crossan, G. P. DNA cross-link repair safeguards genomic stability during premeiotic germ cell development. Nat. Genet. 51, 1283–1294 (2019).
Oberbeck, N. et al. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol. Cell 55, 807–817 (2014).
McWhir, J., Selfridge, J., Harrison, D. J., Squires, S. & Melton, D. W. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5, 217–224 (1993).
Niedernhofer, L. J. et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043 (2006).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Coufal, N. G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009).
Garcia-Perez, J. L. et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–773 (2010).
Kubo, S. et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA 103, 8036–8041 (2006).
Ostertag, E. M., Prak, E. T., DeBerardinis, R. J., Moran, J. V. & Kazazian, H. H. Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000).
Chen, J. M., Stenson, P. D., Cooper, D. N. & Ferec, C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum. Genet. 117, 411–427 (2005).
Kano, H. et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 (2009).
Percharde, M., Sultana, T. & Ramalho-Santos, M. What doesn’t kill you makes you stronger: transposons as dual players in chromatin regulation and genomic variation. BioEssays 42, 1900232 (2020).
Kohlrausch, F. B., Berteli, T. S., Wang, F., Navarro, P. A. & Keefe, D. L. Control of LINE-1 expression maintains genome integrity in germline and early embryo development. Reprod. Sci. 29, 328–340 (2022).
Lewandoski, M., Wassarman, K. M. & Martin, G. R. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148–151 (1997).
de Vries, W. N. et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26, 110–112 (2000).
Protasova, M. S., Andreeva, T. V. & Rogaev, E. I. Factors regulating the activity of LINE1 retrotransposons. Genes 12, 1562 (2021).
Miyoshi, T., Makino, T. & Moran, J. V. Poly(ADP-Ribose) polymerase 2 recruits Replication Protein A to sites of LINE-1 integration to facilitate retrotransposition. Mol. Cell 75, 1286–1298.e12 (2019).
Tristan-Ramos, P. et al. sRNA/L1 retrotransposition: using siRNAs and miRNAs to expand the applications of the cell culture-based LINE-1 retrotransposition assay. Phil. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190346 (2020).
Feng, Q., Moran, J. V., Kazazian, H. H. Jr. & Boeke, J. D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996).
Alter, B. P. Cancer in Fanconi anemia, 1927–2001. Cancer 97, 425–440 (2003).
Rodriguez-Martin, B. et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 52, 306–319 (2020).
Rodić, N. et al. Long Interspersed Element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286 (2014).
Webster, A. L. H. et al. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer. Nature 612, 495–502 (2022).
An, W. et al. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl Acad. Sci. USA 103, 18662–18667 (2006).
Ribeiro, J. & Crossan, G. P. GCNA is a histone binding protein required for spermatogonial stem cell maintenance. Nucleic Acids Res. 51, 4791–4813 (2023).
Pace, P. et al. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21, 3414–3423 (2002).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Acknowledgements
We thank M. Grompe for Fancd2-deficient mice. We thank W. An for the gift of SN1 LINE-1 mice. We thank J. Wysocka for the kind gift of K562 cells carrying the LINE-1 reporter. We thank J. V. Moran for providing the pJJ101/L1.3 and pJJ101/L1.3(D702A) plasmids. We thank K. Burns for the kind gift of the LINE-1 ORF2p antibody. We thank H. Lans and W. Vermeulen for XPF and XPF(D715A) cDNAs. We thank P. Alcón and L. A. Passmore for FANCA and FANCC cDNAs. We thank the Fanconi Anemia Research Foundation for providing the PD20, PD220 and PD331 cell lines. We thank the Human Research Tissue Bank (National Institute for Health Research Cambridge Biomedical Research Centre) for processing the histology. We thank M. Hodskinson for purifying recombinant proteins and assisting in biochemical experiments. We thank L. Mulderrig for his assistance in performing in vitro retrotransposition assays and F. Langevin for his assistance in generating K562 knockout cell lines. We thank R. Hill for his assistance with embryo dissection, primordial germ cell quantification, histology and ploidy assays. We are grateful to J. Clark, D. Karpinski, M. Wiacek, C. Knox and Ares and Biomedical Staff for their help with mouse experimental work. We thank M. Daly, P. Andrée Penttilä, F. Zhang and the Flow Cytometry Core staff for their technical assistance. N.B. is supported by the César Milstein Studentship from the Darwin Trust of Edinburgh. This work was supported by the Medical Research Council as part of UK Research and Innovation (file reference no. MC_UP_1201/18).
Author information
Authors and Affiliations
Contributions
N.B. and G.P.C. conceived the study and wrote the manuscript. N.B. conducted all experiments and analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Zsuzsanna Izsvak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Carolina Perdigoto, Tiago Faial and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Fanconi anemia DNA crosslink repair factors restrict endonuclease-independent integration events.
a) Schema illustrating the L1(D205A)-G418R retrotransposition assay in K562 cells. ORF2 endonuclease activity was disrupted by a point mutation (D205A) generated by site-directed mutagenesis. Top, immunoblot for Cas9 in K562 cells. Bottom, PCR validating integration of the L1(D205A)-G418R reporter (both ends of the construct) in genomic DNA extracted from the K562 cells expressing Cas9 (representative data from 2 independent experiments). b) K562 cells carrying the endonuclease dead L1(D205A)-G418R (EN-) reporter were treated with DOX and then plated into selective methylcellulose media (containing 1 mg/mL G418). Colonies were quantified after 10 days. Each dot corresponds to an independent experiment. c-f) DNA repair-deficient mutants on the L1(D205A)-G418R (EN-) background were assayed. Each dot corresponds to an independent experiment: c) Homologous recombination (HR), d) Translesion synthesis (TLS), e) Interstrand crosslink (ICL) repair and f) Nucleases. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by two-tailed Mann–Whitney U-test.
Extended Data Fig. 2 DNA damage following DOX induction in K562 wildtype cell line.
a) Representative images of γ-H2A.x foci in wildtype or ΔFANCD2 K562 cells (that do not contain the L1-G418R reporter) before and after DOX treatment for 48 hours. Scale bar, 10 μm. b) Quantification of the percentage of wildtype or ΔFANCD2 K562 cells with >5 foci, n = 3 independent experiments, the bar represents the mean, the error bars represent the s.e.m. and P values were calculated by two-tailed Mann-Whitney U-test.
Extended Data Fig. 3 Fluctuation analysis in K562 L1-G418R cell-lines, LINE-1 protein expression in mutant cell lines, and cell fitness in complemented human Fanconi fibroblasts.
Independent cultures of each mutant were established from single cells and treated with DOX for 10 days. Each culture was then plated in one well of a 6-well plate containing semi-solid methylcellulose media and G418. Plating efficiency was determined for four cultures by performing three 10-fold serial dilutions and plating without G418. Statistical analysis was performed as described previously46. n = 3 independent experiments, the dot represents the mean and the error bars represent the s.e.m., P values were calculated by two-tailed Mann-Whitney U-test; a) SLX4, XPF and ERCC1 (SXE), b) Fanconi anemia (FA), c) Homologous recombination (HR), d) Translation synthesis (TLS), e) Nucleotide excision repair (NER), f) Non-homologous end joining (NHEJ) and g) Bloom syndrome helicase (BLM). h) Immunoblot for histone H3, LINE-1 ORF1p and LINE-1 ORF2p in wildtype, ΔFANCA, ΔFANCD2 and ΔXPF K562 L1-G418R cells after DOX treatment; H3 loading control (representative data from 2 independent experiments). i-k) P220 (ΔFANCA) and PD331 (ΔFANCC) human fibroblasts were complemented by ectopically expressing FANCA and FANCC cDNA, respectively. The cellular sensitivity of these cell lines to mitomycin C was tested and compared with the previously complemented cell line PD20 (ΔFANCD2); i) PD20 and PD20 complemented cell lines, j) PD220 and PD220 complemented cell lines, k) PD331 and PD331 complemented cell lines (data in i-k represent mean and s.e.m.; n = 3 independent experiments).
Extended Data Fig. 4 SLX4 and XPF complementation and double-knockout mutants.
a) Immunoblot in clones targeted for XPF with sgRNA1 in wildtype or mutant K562 L1-G418R; TUBULIN, α-tubulin loading control. b) The cellular sensitivity of the K562 L1-G418R XPF mutants to mitomycin C (n = 3 independent experiments). c) Schema depicting XPF-ERCC1 structure-specific endonuclease (top) and XPF-ERCC1 catalytically dead due to the D715A point mutation (bottom). d) Immunoblot for XPF in ΔXPF#13 clone ectopically expressing either XPF or XPF(D715A) mutant cDNA. Tubulin loading control. e) The cellular sensitivity of the K562 L1-G418R XPF mutant and complemented cell lines to mitomycin C (n = 3 independent experiments). f) L1 retrotransposition measurement using L1-G418R in K562 cells (wildtype, XPF mutant and complementation cell lines), each point corresponds to an independent experiment. g) Immunoblot in clones targeted for ERCC1 in wildtype K562 L1-G418R; TUBULIN, α-tubulin loading control. h) Immunoblot in clones targeted for ERCC1 in K562 L1-G418R ΔXPF (sg1) cell line; TUBULIN, α-tubulin loading control. i) The cellular sensitivity of the K562 L1-G418R ERCC1 mutant to mitomycin C (n = 3 independent experiments). j) L1 retrotransposition measurement using L1-G418R in K562 cells (wildtype, ERCC1, XPF and double-knockout cell lines), each point corresponds to an independent experiment. k) Immunoblot for FLAG in ΔSLX4 clones ectopically expressing either: right, mini-SLX4 (amino acids 1–750) or left, SLX4-FL (amino acids 1-1834) cDNA. l) The cellular sensitivity of the SLX4 mutant and complemented cell lines to mitomycin C (n = 3 independent experiments). Data represent mean and s.e.m. P values were calculated by two-tailed Mann-Whitney U-test.
Extended Data Fig. 5 SLX4 potentiates the nuclease activity of XPF-ERCC1 on DNA:DNA and DNA:RNA substrates.
a) Silver staining of the purified XE and SXE complexes used in the previous biochemistry assays (representative data from 2 independent experiments). b) Schema illustrating the substrate (annealed oligonucleotides) used in nuclease assays without a gap in the junction. c) Schema illustrating the substrate (annealed oligonucleotides) for nuclease assays with a gap in the junction (2, 5 or 9 nucleotides). XE and SXE were reacted with different fluorescent-labelled nucleic acid substrates, over a time course (0.2, 5, 10, 20, and 60 minutes). The reaction products were separated by 12% denaturing PAGE (top panel), and the decay of the substrate band was quantified and expressed as a percentage of the initial substrate; data were fitted using single exponential decay (bottom panel, left) in order to calculate reaction rates (bottom panel, right). XE data are plotted in blue; SXE data are plotted in red, n = 2 independent experiments, error bars represent s.e.m. A gap of d) 0, e) 2 or f) 5 nucleotides was introduced between the junction of splayed arms and the annealed DNA. A gap of g) 0, h) 2 or i) 9 nucleotides was introduced between the junction of splayed arms and the annealed RNA.
Extended Data Fig. 6 Loss of cGAS does not rescue the Ercc1−/− phenotype.
a) Left, schema illustrating the treatment regime in K562 L1-G418R cells to assess if L1 expression induces an interferon response. Right, the expression of IFN and IFN-induced genes was assessed by RT-qPCR. n = 2 cultures per condition. Expression was normalized to Gapdh and made relative to the untreated sample. b) The expression of Ifna, Cgas and Sting was assessed by RT-qPCR. Each point represents one mouse. Expression was normalized to Gapdh and made relative to the untreated sample. c) Survival curve of wildtype, Ercc1−/− and Ercc1−/−Cgas−/− mice. No statistically significant difference (P = 0.0084) was found with the Wilcoxon test between Ercc1−/− and Ercc1−/−Cgas−/− mice. d) Weight curve Ercc1−/− and Ercc1−/−Cgas−/− mice (n = 3 per mice group). Representative histological analysis of; e) liver (scale bar: 40 μm) and f) kidney (scale bar: 20 μm) of Ercc1−/−Cgas−/− and littermate controls. g) Frequency of 16n hepatocytes quantified by flow cytometry, each point represents one animal. h) Frequency of 16n kidney cells quantified by flow cytometry, each point represents one animal i, j) Frequency of SSEA1+ primordial germ cells in Ercc1−/−Cgas−/− and littermate controls at E12.5 quantified by flow cytometry, each point represents one embryonic gonad. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by two-tailed Mann-Whitney U-test.
Extended Data Fig. 7 FA DNA ICL repair factors restrain SN1 LINE-1 retrotransposition.
Quantification of the frequency of SN1 integrations by ddPCR in adult mice. Each point represents one mouse. Various tissues with different replication potentials were assessed in; a) Ercc1−/−, b) Fancd2−/−, c) Fancd2−/R, or d) Fanca−/− adult mice. e) Schema illustrating the timing of peripheral blood harvests from wildtype and Fanca−/− adult mice. f) Quantification of the frequency of SN1 integrations in peripheral white blood cells from the same wildtype and Fanca−/− mice by ddPCR at 3, 6 and 9 months. n = 2 independent mice per genotype. g) The expression of SN1 LINE-1 reporter was quantified by RT-qPCR in peripheral white blood cells from adult mice at 6 and 9 months. Expression was normalized to Gapdh and made relative to the 9 months sample. n = 3 independent mice per genotype. h) Same analysis but including E18.5 liver sample. n = 3 independent mice per genotype. i) Representative histological images of the testes of wildtype or Fanca−/− adult mice. Scale bar, 200 μm. j) Quantification of the frequency of tubules exhibiting spermatogenesis. Each point represents one mouse k, l) Quantification of the frequency of SN1 integrations in; k) testis from adult wildtype and Fancd2−/− mice and l) fetal testis from wildtype and Ercc1−/− E18.5 embryos by ddPCR. Each point represents one mouse or embryo. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by two-tailed Mann-Whitney U-test.
Extended Data Fig. 8 SN1 LINE-1 retrotransposition in wildtype and Fanconi mice analysed by FACS.
Representative flow cytometry plot and quantification of GFP+ cells from SN1, SN1 Fanca−/− and SN1 Fancd2−/− mice from a) kidney, b) bone marrow, c) lung, d) testis. Each point represents one mouse, data represent mean and s.e.m. P values were calculated by two-tailed Mann-Whitney U-test e) Representative flow cytometry plot of single-cell suspension derived from bone marrow and testis of wildtype mice (that did not carry the SN1 LINE-1 reporter).
Extended Data Fig. 9 FA DNA ICL repair factors restrain SN1 LINE-1 retrotransposition during embryogenesis.
Quantification of the frequency of SN1 integrations was quantified by ddPCR in E18.5 embryos. Each point represents one embryo. Various tissues with different replicative potentials were assessed in; a) Ercc1−/−, b) Fanca−/−, c) Fancd2−/−, or d) Fancd2-/R E18.5 embryos. e) The expression of SN1 LINE-1 reporter (Sn1 Line-1 Orf2p) was quantified by RT-qPCR in multiple tissues of wildtype and Ercc1−/− adult mice and embryos carrying the SN1 LINE-1 reporter. Expression was normalized to Gapdh and made relative to wildtype E18.5 embryos. n = 3 adult mice per genotype and n = 3 embryos per genotype f) The expression of SN1 LINE-1 reporter (Sn1 Line-1 Orf2p) was quantified by RT-qPCR in multiple tissues of wildtype adult mice and embryos carrying the SN1 LINE-1 reporter. Expression was normalized to Gapdh and made relative to the adult heart. Each point represents one animal. Data represent mean and s.e.m. P values were calculated by two-tailed Mann-Whitney U-test.
Extended Data Fig. 10 Maternal and zygotic ERCC1 contribute to LINE-1 retrotransposition restriction.
a) Schema showing the paternal genotype, the maternal Ercc1 genotype and the inheritance of ERCC1 in the oocyte/zygote. In this cross, maternal Ercc1 mRNA is present in the oocyte. E18.5 embryos were obtained and frequency of spliced eGFP in multiple tissues was assessed by ddPCR. Each point represents one embryo. b) In this cross, maternal Ercc1 mRNA is absent in the oocyte. E18.5 embryos were obtained and frequency of spliced eGFP in multiple tissues was assessed by ddPCR. Each point represents one embryo. c) In this cross, maternal Ercc1 mRNA is present in the oocyte. E18.5 embryos were obtained and the frequency of SN1 integration in multiple tissues was assessed by ddPCR. Frequency of spliced eGFP was compared between Ercc1−/− (from Ercc1+/− intercrosses) and Ercc1Δ/Δ embryos from Ercc1+/Δ intercrosses. Each point represents one embryo. Table showing the frequency of Ercc1−/− was as expected at E18.5. The P value was calculated by two-sided Fischer’s exact test. d) In this cross, maternal Ercc1 mRNA is present in the oocyte. E18.5 embryos were obtained and spliced eGFP frequency in multiple tissues was assessed by ddPCR. Each point represents one embryo. Table showing the frequency of Ercc1−/− was as expected at E18.5. The P value was calculated by two-sided Fischer’s exact test. Data represent mean and s.e.m. Unless otherwise specified, P values were calculated by two-tailed Mann-Whitney U-test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.
Supplementary Tables
Supplementary Tables 1–3.
Supplementary Data
Source data for Supplementary Figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bona, N., Crossan, G.P. Fanconi anemia DNA crosslink repair factors protect against LINE-1 retrotransposition during mouse development. Nat Struct Mol Biol 30, 1434–1445 (2023). https://doi.org/10.1038/s41594-023-01067-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01067-8