Abstract
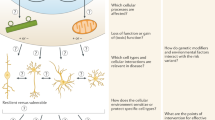
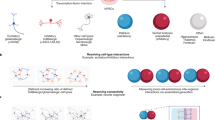
Complex brain disorders are highly heritable and arise from a complex polygenic risk architecture. Many disease-associated loci are found in non-coding regions that house regulatory elements. These elements influence the transcription of target genes—many of which demonstrate cell-type-specific expression patterns—and thereby affect phenotypically relevant molecular pathways. Thus, cell-type-specificity must be considered when prioritizing candidate risk loci, variants and target genes. This Review discusses the use of high-throughput assays in human induced pluripotent stem cell-based neurodevelopmental models to probe genetic risk in a cell-type- and patient-specific manner. The application of massively parallel reporter assays in human induced pluripotent stem cells can characterize the human regulome and test the transcriptional responses of putative regulatory elements. Parallel CRISPR-based screens can further functionally dissect this genetic regulatory architecture. The integration of these emerging technologies could decode genetic risk into medically actionable information, thereby improving genetic diagnosis and identifying novel points of therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sims, R. et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1384 (2017).
Curran, S., Ahn, J. W., Grayton, H., Collier, D. A. & Ogilvie, C. M. NRXN1 deletions identified by array comparative genome hybridisation in a clinical case series - further understanding of the relevance of NRXN1 to neurodevelopmental disorders. J. Mol. Psychiatry 1, 4 (2013).
Betancur, C. & Buxbaum, J. D. SHANK3 haploinsufficiency: a “common” but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol. Autism 4, 17 (2013).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke, S., Walters, J.T.R & O’Donovan, M.C. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. Preprint at medRxiv https://doi.org/10.1101/2020.09.12.20192922 (2020).
Arnold, P. D. et al. International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 51, 1207–1214 (2019).
Mullins, N. et al. Genome-wide association study of over 40,000 bipolar disorder cases provides novel biological insights. Preprint at medRxiv https://doi.org/10.1101/2020.09.17.20187054 (2020).
Hettema, J. M. et al. Genome-wide association study of shared liability to anxiety disorders in Army STARRS. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 183, 197–207 (2020).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Yu, D. et al. Interrogating the genetic determinants of Tourette’s syndrome and other tiC disorders through genome-wide association studies. Am. J. Psychiatry 176, 217–227 (2019).
Duncan, L. E. et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry 23, 666–673 (2018).
Huckins, L. M. et al. Analysis of genetically regulated gene expression identifies a prefrontal PTSD gene, SNRNP35, specific to military cohorts. cell rep. 31, 107716 (2020).
Lee, P. H. et al. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders.Cell 179, 1469–1482.e11 (2019).
Montalbano, A., Canver, M. C. & Sanjana, N. E. High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell 68, 44–59 (2017).
Edwards, S. L., Beesley, J., French, J. D. & Dunning, A. M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 (2013).
Fernando, M. B., Ahfeldt, T. & Brennand, K. J. Modeling the complex genetic architectures of brain disease. Nat. Genet. 52, 363–369 (2020).
van Hugte, E. & Nadif Kasri, N. Modeling psychiatric diseases with induced pluripotent stem cells. in Frontiers in Psychiatry. Advances in Experimental Medicine and Biology vol. 1192 (ed. Kim, Y.-K.) 297–312 (Springer, 2019).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Albert, F. W. & Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16, 197–212 (2015).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Wallace, C. Statistical testing of shared genetic control for potentially related traits. Genet. Epidemiol. 37, 802–813 (2013).
Plagnol, V., Smyth, D. J., Todd, J. A. & Clayton, D. G. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics 10, 327–334 (2009).
Wen, X., Pique-Regi, R. & Luca, F. Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS Genet. 13, e1006646 (2017).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Giambartolomei, C. et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 34, 2538–2545 (2018).
Dobbyn, A. et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 102, 1169–1184 (2018). This study demonstrates the ability of colocalization analyses to identify candidate SNPs that are predictive of disease-associated gene expression in a tissue-specific manner. Identification of the FURIN SNP by this method was functionally validated in ref. 68.
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 (2019).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Barbeira, A.N., Dickinson, S.P. & Bonazzola, R. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Park, Y. et al. A Bayesian approach to mediation analysis predicts 206 causal target genes in Alzheimer’s disease. Preprint at bioRxiv https://doi.org/10.1101/219428 (2017).
Fromer, M., Roussos, P. & Sieberts, S. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Supplementary materials. Science 369, 1318–1330 (2020).
Kim-Hellmuth, S. et al. Cell type-specific genetic regulation of gene expression across human tissues. Science 369, eaaz8528 (2020). One of the most recent publications from the GTEx Consortium describes their efforts to map QTLs to human tissues. More importantly, it reports the use of 43 pairs of tissues and cell types to identify cell-type-specific genetic regulation of gene expression.
Donovan, M. K. R., D’Antonio-Chronowska, A., D’Antonio, M. & Frazer, K. A. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat. Commun. 11, 955 (2020).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Huckins, L. M. et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 51, 659–674 (2019).
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Sey, N.Y.A., Hu, B. & Mah, W. et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 23, 583–593 (2020).
Gusev, A. et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95, 535–552 (2014).
Gasperini, M. et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 377–390.e19 (2019). A prime example of a CRISPR perturbation screen design based on eQTLs overlaping with GWAS loci. A framework was later described in depth as crispQTL by the same group.
Nott, A. et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019).
Xie, S., Duan, J., Li, B., Zhou, P. & Hon, G. C. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017).
Skene, N. G. & Grant, S. G. N. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16 (2016).
Ho, S. M. et al. Evaluating synthetic activation and repression of neuropsychiatric-related genes in hiPSC-derived NPCs, neurons, and astrocytes. Stem Cell Reports 9, 615–628 (2017).
Duan, J., Penzes, P. & Gejman, P. From genetic association to disease biology: 2D and 3D human IPSC models of neuropsychiatric disorders and CRISPR/Cas9 genome editing. Eur. Neuropsychopharmacol. 29, S763–S764 (2019). Suppl. 3.
Jerber, J. et al. Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Preprint at bioRxiv https://doi.org/10.1101/2020.05.21.103820 (2020).
Neavin, D. et al. Single cell eQTL analysis identifies cell type-specific genetic control of gene expression in fibroblasts and reprogrammed induced pluripotent stem cells. Preprint at bioRxiv https://doi.org/10.1101/2020.06.21.163766 (2020).
Mitchell, J. M. et al. Mapping genetic effects on cellular phenotypes with ‘cell villages’. Preprint at bioRxiv https://doi.org/10.1101/2020.06.29.174383 (2020).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Rajarajan, P. et al. Spatial genome exploration in the context of cognitive and neurological disease. Curr. Opin. Neurobiol. 59, 112–119 (2019).
Rehbach, K., Fernando, M. B. & Brennand, K. J. Integrating CRISPR Engineering and hiPSC-Derived 2D Disease Modeling Systems. J. Neurosci. 40, 1176–1185 (2020).
Nehme, R. & Barrett, L. E. Using human pluripotent stem cell models to study autism in the era of big data. Mol. Autism 11, 21 (2020).
Deneault, E. et al. Complete disruption of autism-susceptibility genes by gene editing predominantly reduces functional connectivity of isogenic human neurons. Stem Cell Reports 11, 1211–1225 (2018).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Larson, M. H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013).
Hilton, I. B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Balboa, D. et al. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Reports 5, 448–459 (2015).
Savell, K. E. et al. A neuron-optimized CRISPR/dCas9 activation system for robust and specific gene regulation. eNeuro 6, 1–17 (2019).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-Derived neurons. Neuron 104, 239–255.e12 (2019).
Schrode, N. et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 51, 1475–1485 (2019). This paper nicely shows how the colocalization of GWAS and eQTL data, used to identify a putative causal variant (FURIN) in ref. 31, can be functionally validated in vitro. Specifically, this study used CRISPR editing and allelic conversion of a causal allele in hiPSC-derived neurons to measure phenotypic effects.
Nickolls, A. R. et al. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 30, 932–946.e7 (2020).
Lalli, M. A., Avey, D., Dougherty, J. D., Milbrandt, J. & Mitra, R. D. High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation. Genome Res. 30, 1317–1331 (2020).
Zaslavsky, K. et al. SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat. Neurosci. 22, 556–564 (2019).
Zhang, S. et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science 369, 561–565 (2020). This work identifies allele-specific and disorder-associated open chromatin regions in hiPSC neurons. This study also identified a 26–29% overlap between allele-specific open chromatin regions and non-neuronal MPRA SNPs from ref. 86. The ATAC–seq datasets from this study could be integrated with information from future or existing MPRA datasets to provide endogenous context and match regulatory enhancer activity to specific brain cell types.
Breen, M.S., Browne, A. & Hoffman, G.E. et al. Transcriptional signatures of participant-derived neural progenitor cells and neurons implicate altered Wnt signaling in Phelan-McDermid syndrome and autism. Molecular Autism 11, 53 (2020).
de Boer, C. G. et al. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol. 38, 56–65 (2020). This paper reports one of the largest MPRA libraries, assessing the regulatory logic of 100 million promoters in vivo. Although performed in yeast, this study hints at the possibility of increasing the scale of MPRAs in other cell types.
Birnbaum, R. Y. et al. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet. 10, e1004592 (2014).
Patwardhan, R. P. et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 27, 1173–1175 (2009).
Kheradpour, P. et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 23, 800–811 (2013).
Gordon, M. G. et al. lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nat. Protoc. 15, 2387–2412 (2020).
van Arensbergen, J. et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet. 51, 1160–1169 (2019). This work describes a novel MPRA technique, SuRE MPRA, that allows for substantial expansion of library size. By creating two random genome-expanding libraries containing ~300 million variants, this study identifies transcriptional shifts for 5.9 million SNPs in human cells and provides a dataset that can assist in the identification of casual alleles from the list of eQTL and GWAS hits.
Fulco, C. P. et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). This paper describes the CRISPRi-FlowFISH method, a CRISPR screening appraoch using scRNA-seq. Although it has limited scalability, fluorescence-activated cell sorting and scRNA-seq can expand the application and resolution of this technique. By developing an activity-by-contact model of regulatory logic, this study integrates CRISPR screens, scRNA-seq and ATAC–seq data.
Kircher, M. et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nat. Commun. 10, 3583 (2019).
Inoue, F., Kreimer, A., Ashuach, T., Ahituv, N. & Yosef, N. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell 25, 713–727.e10 (2019). This study expands the application of past MPRAs by using a lentiviral technique for chromosomal integration. To our knowledge, this study is the first instance of MPRAs in hiPSC neural progenitor cells, thereby showing the potential for massively parallel assays to be adapted for use in hiPSC models to provide cell-specific and donor-specific context.
Melnikov, A. et al. Massively parallel reporter assays in cultured mammalian cells. J. Vis. Exp. 90, 51719 (2014).
Inoue, F. et al. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 27, 38–52 (2017).
Uebbing, S. et al. Massively parallel discovery of human-specific substitutions that alter neurodevelopmental enhancer activity. Preprint at bioRxiv https://doi.org/10.1101/865519 (2019). To our knowledge, is the first report of MPRAs in human-derived neurons. While previous studies successfully adapted MPRAs for use in human-derived neural stem cells, this study successfully probed thousands of enhancers associated with human corticogenesis and neurodevelopment.
Tewhey, R. et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016).
Ulirsch, J. C. et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell 165, 1530–1545 (2016).
Myint, L. et al. A screen of 1,049 schizophrenia and 30 Alzheimer’s-associated variants for regulatory potential. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 183, 61–73 (2020).
Geller, E. et al. Massively parallel disruption of enhancers active during human corticogenesis. bioRxiv https://doi.org/10.1101/852673 (2019).
Nicholas, C. R. et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 12, 573–586 (2013).
Tian, A., Muffat, J. & Li, Y. Studying human neurodevelopment and diseases using 3D brain organoids. J. Neurosci. 40, 1186–1193 (2020).
Burke, E. E. et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 11, 462 (2020).
Klein, J.C. et al. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nat. Methods https://doi.org/10.1038/s41592-020-0965-y (2020).
Ernst, J. et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat. Biotechnol. 34, 1180–1190 (2016).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Diao, Y. et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14, 629–635 (2017).
Jin, X. et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with Autism risk genes. Preprint at bioRxiv https://doi.org/10.1101/791525 (2019). This work applies the Perturb-seq technique first reported in ref. 95 to interrogate the influence of risk genes for a neurodevelopmental disorder in an hiPSC model. Phenotypic aberrations in neuronal and glial cells support the use of CRISPR-based screens with scRNA-seq to determine cell-type-specific mechanisms underlying risk for complex brain disorders.
Gasperini, M., Tome, J. M. & Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 21, 292–310 (2020).
Mimitou, E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019).
Cederquist, G. Y. et al. A multiplex human pluripotent stem cell platform defines molecular and functional subclasses of autism-related genes. Cell Stem Cell 27, 35–49.e6 (2020).
Tian, R. et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Preprint at bioRxiv https://doi.org/10.1101/2020.06.27.175679 (2020). This study used three screening techniques (genome-wide random screens, a CRISPRa/i target screen and CRISPR droplet sequencing (CROP-seq)) to conduct pooled CRISPR screens with single-cell transcriptome resolution for the identification and further exploration of oxidative-stress related enhancers in human neurons. This study provides evidence for the application of complex and large-scale CRISPRa/i screens in mature human brain cells.
Liu, Y. et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23, 758–771.e8 (2018).
Yang, J. et al. Genome-scale CRISPRa screen identifies novel factors for cellular reprogramming. Stem Cell Reports 12, 757–771 (2019).
Gaffney, D. J. Mapping and predicting gene-enhancer interactions. Nat. Genet. 51, 1662–1663 (2019).
Abud, E. M. et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293.e9 (2017).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Bliss, L. A. et al. Use of postmortem human dura mater and scalp for deriving human fibroblast cultures. PLoS One 7, e45282 (2012).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Theka, I. et al. Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. Stem Cells Transl. Med. 2, 473–479 (2013).
Barretto, N. et al. ASCL1- and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. J. Neurosci. Methods 334, 108548 (2020).
Yang, N. et al. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14, 621–628 (2017).
Vadodaria, K. C. et al. Generation of functional human serotonergic neurons from fibroblasts. Mol. Psychiatry 21, 49–61 (2016).
Tcw, J. et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports 9, 600–614 (2017).
Ehrlich, M. et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc. Natl. Acad. Sci. USA 114, E2243–E2252 (2017).
Li, Y. et al. Development of human in vitro brain-blood barrier model from induced pluripotent stem cell-derived endothelial cells to predict the in vivo permeability of drugs. Neurosci. Bull. 35, 996–1010 (2019).
Marton, R. M. et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 22, 484–491 (2019).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Hoffman, G. E. et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 8, 2225 (2017).
Kathuria, A. et al. Transcriptomic landscape and functional characterization of induced pluripotent stem cell-derived cerebral organoids in schizophrenia. JAMA Psychiatry 77, 745–754 (2020).
Acknowledgements
This work was partially supported by National Institute of Health (NIH) grants R56 MH101454 (K.J.B), R01 MH106056 (K.J.B.), R01 MH109897 (K.J.B.) and R01 MH118278 (L.M.H.). All figures in this review were created with BioRender.com
Author information
Authors and Affiliations
Contributions
K.G.T., K.J.B., and L.M.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Peer review information Nature Neuroscience thanks Martin Kampmann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Townsley, K.G., Brennand, K.J. & Huckins, L.M. Massively parallel techniques for cataloguing the regulome of the human brain. Nat Neurosci 23, 1509–1521 (2020). https://doi.org/10.1038/s41593-020-00740-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00740-1
This article is cited by
-
Frequency, morbidity and equity — the case for increased research on male fertility
Nature Reviews Urology (2024)
-
Integrating genetics and transcriptomics to study major depressive disorder: a conceptual framework, bioinformatic approaches, and recent findings
Translational Psychiatry (2023)
-
Probing neural circuit mechanisms in Alzheimer’s disease using novel technologies
Molecular Psychiatry (2023)
-
Comprehensive and integrative analyses identify TYW5 as a schizophrenia risk gene
BMC Medicine (2022)
-
What next for eating disorder genetics? Replacing myths with facts to sharpen our understanding
Molecular Psychiatry (2022)