Abstract
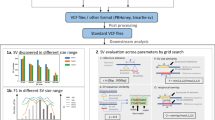
Complete, telomere-to-telomere (T2T) genome assemblies promise improved analyses and the discovery of new variants, but many essential genomic resources remain associated with older reference genomes. Thus, there is a need to translate genomic features and read alignments between references. Here we describe a method called levioSAM2 that performs fast and accurate lift-over between assemblies using a whole-genome map. In addition to enabling the use of several references, we demonstrate that aligning reads to a high-quality reference (for example, T2T-CHM13) and lifting to an older reference (for example, Genome reference Consortium (GRC)h38) improves the accuracy of the resulting variant calls on the old reference. By leveraging the quality improvements of T2T-CHM13, levioSAM2 reduces small and structural variant calling errors compared with GRC-based mapping using real short- and long-read datasets. Performance is especially improved for a set of complex medically relevant genes, where the GRC references are lower quality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The Illumina data are from Baid et al.37. The PacBio-HiFi data are from Jarvis et al.41. The HG002 ONT data were sequenced at the Human Genome Sequencing Center, Baylor College of Medicine, and are available at https://www.ncbi.nlm.nih.gov/sra/PRJNA930475. Source data are provided with this paper.
Code availability
The software is available at https://github.com/milkschen/leviosam2 under the MIT license64. The experiments described in this paper are further described at https://github.com/milkschen/levioSAM2-experiments under the MIT license65.
References
Schneider, V. A. et al. Evaluation of grch38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Guo, Y. et al. Improvements and impacts of grch38 human reference on high throughput sequencing data analysis. Genomics 109, 83–90 (2017).
Aganezov, S. et al. A complete reference genome improves analysis of human genetic variation. Science 376, eabl3533 (2022).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Smigielski, E. M., Sirotkin, K., Ward, M. & Sherry, S. T. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 28, 352–355 (2000).
Mailman, M. D. et al. The NCBI dbGAP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature 590, 290–299 (2021).
Consortium, G. The GTEX Consortium Atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Lowy-Gallego, E. et al. Variant calling on the GRCh38 assembly with the data from phase three of the 1000 genomes project. Wellcome Open Res. 4, 50 (2019).
Salzberg, S. L. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 20, 92 (2019).
Gao, G. F. et al. Before and after: comparison of legacy and harmonized TCGA genomic data commons’ data. Cell Syst. 9, 24–34 (2019).
Lansdon, L. A. et al. Factors affecting migration to GRCh38 in laboratories performing clinical next-generation sequencing. J. Mol. Diagn. 23, 651–657 (2021).
Fujita, P. A. et al. The UCSC genome browser database: update 2011. Nucleic Acids Res. 39, D876–D882 (2010).
Zhao, H. et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014).
Picard toolkit. GitHub https://broadinstitute.github.io/picard/ (2019).
Mun, T., Chen, N.-C. & Langmead, B. Leviosam: fast lift-over of variant-aware reference alignments. Bioinformatics 37, 4243–4245 (2021).
Pan, B. et al. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinformatics 20, 17–29 (2019).
Ormond, C., Ryan, N. M., Corvin, A. & Heron, E. A. Converting single nucleotide variants between genome builds: from cautionary tale to solution. Brief. Bioinform. 22, bbab069 (2021).
Li, H. et al. Exome variant discrepancies due to reference genome differences. Am. J. Hum. Genet. 108, 1239–1250 (2021).
Lansdon, L. A. et al. Clinical validation of genome reference consortium human build 38 in a laboratory utilizing next-generation sequencing technologies. Clin. Chem. 68, 1177–1183 (2022).
Behera, S. et al. FixItFelix: improving genomic analysis by fixing reference errors. Genome Biol. 24, 31 (2023).
Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics 37, 1639–1643 (2021).
Chen, N.-C., Solomon, B., Mun, T., Iyer, S. & Langmead, B. Reference flow: reducing reference bias using multiple population genomes. Genome Biol. 22, 8 (2021).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Preprint at bioRxiv https://doi.org/10.1101/201178 (2018).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Wagner, J. et al. Benchmarking challenging small variants with linked and long reads. Cell Genom. 2, 100128 (2022).
Wagner, J. et al. Curated variation benchmarks for challenging medically relevant autosomal genes. Nat. Biotechnol. 40, 672–680 (2022).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019).
Zook, J. M. et al. A robust benchmark for detection of germline large deletions and insertions. Nat. Biotechnol. 38, 1347–1355 (2020).
Holtgrewe, M. Mason: A Read Simulator for Second Generation Sequencing Data. Report No. TR-B-10-06 (Technical Reports of Institut für Mathematik und Informatik, Freie Universität Berlin, 2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
Baid, G. et al. An extensive sequence dataset of gold-standard samples for benchmarking and development. Preprint at bioRxiv https://doi.org/10.1101/2020.12.11.422022 (2020).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Jain, C., Rhie, A., Hansen, N. F., Koren, S. & Phillippy, A. M. Long-read mapping to repetitive reference sequences using winnowmap2. Nat. Methods 19, 705–710 (2022).
Jarvis, E. D. et al. Semi-automated assembly of high-quality diploid human reference genomes. Nature 611, 519–531 (2022).
Smolka, M. et al. Comprehensive structural variant detection: from mosaic to population-level. Preprint at bioRxiv https://doi.org/10.1101/2022.04.04.487055 (2022).
English, A. C., Menon, V. K., Gibbs, R. A., Metcalf, G. A. & Sedlazeck, F. J. Truvari: refined structural variant comparison preserves allelic diversity. Genome Biol. 23, 271 (2022).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Mandelker, D. et al. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet. Med. 18, 1282–1289 (2016).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Talenti, A. & Prendergast, J. nf-LO: a scalable, containerized workflow for genome-to-genome lift over. Genome Biol. Evol. 13, evab183 (2021).
Garrison, E. & Guarracino, A. Unbiased pangenome graphs. Bioinformatics 39, btac743 (2023).
Ebert, P. et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372, eabf7117 (2021).
Delcher, A. L. et al. Alignment of whole genomes. Nucleic Acids Res. 27, 2369–2376 (1999).
Chin, C.-S. et al. Multiscale analysis of pangenomes enables improved representation of genomic diversity for repetitive and clinically relevant genes. Nat. Methods 20, 1213–1221 (2023).
Gog, S., Beller, T., Moffat, A. & Petri, M. From theory to practice: plug and play with succinct data structures. In Proc. 13th International Symposium on Experimental Algorithms (eds. Gudmundsson, J. & Katajainen, J.) 326–337 (SEA, 2014).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Rapid yaml. GitHub https://github.com/biojppm/rapidyaml (2022).
Bonfield, J. K. et al. Htslib: C library for reading/writing high-throughput sequencing data. Gigascience 10, giab007 (2021).
Pockrandt, C., Alzamel, M., Iliopoulos, C. S. & Reinert, K. GenMap: ultra-fast computation of genome mappability. Bioinformatics 36, 3687–3692 (2020).
Leitner-Ankerl, M. Robin hood unordered map and set. GitHub https://github.com/martinus/robin-hood-hashing (2022).
Quinlan, A. R. & Hall, I. M. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Martin, M. et al. Whatshap: fast and accurate read-based phasing. Preprint at bioRxiv https://doi.org/10.1101/085050 (2016).
Cook, D., Kolesnikov, A., Chang, P.-C. & Carroll, A. Improving variant calling using haplotype information. DeepVariant Blog https://google.github.io/deepvariant/posts/2021-02-08-the-haplotype-channel/ (2021).
Krusche, P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 37, 555–560 (2019).
Gordon, A. Gnu time. https://www.gnu.org/software/time/ (2018).
Chen, N.-C. leviosam2. Zenodo https://doi.org/10.5281/zenodo.8198490 (2023).
Chen, N.-C. levioSAM2-experiments v.0.1. Zenodo https://doi.org/10.5281/zenodo.8198541 (2023).
Acknowledgements
We thank T. Mun for his advice and contribution to the levioSAM2 programming infrastructure. We appreciate advice from H.-C. Chen on software deployment, C. Pockrandt on mappability resources, A. Shumate on gene lift-over and S. Zarate on T2T-CHM13 variant analysis. We also thank A. Carroll and P.-C. Chang for DeepVariant discussions, A. Rhie for T2T-CHM13 discussions and J. Zook for GIAB strata suggestions. N.-C.C. and B.L. were supported by National Institutes of Health (NIH) grants R01HG011392 and R35GM139602 to B.L. F.J.S. and L.F.P. were supported by NIH grants 1U01HG011758-01 and UM1HG008898. S.K. and A.M.P. were supported by the Intramural Research Program of the National Human Genome Research Institute (NHGRI), NIH. Part of this research project was conducted using computational resources at the Maryland Advanced Research Computing Center (MARCC). Prebuilt levioSAM2 resources for T2T-CHM13 to GRC references are made freely available on Amazon Web Services thanks to the AWS Public Dataset Program.
Author information
Authors and Affiliations
Contributions
N.-C.C., S.K., A.M.P. and B.L. designed the method. N.-C.C. wrote the software. N.-C.C. and L.F.P. performed the experiment. N.-C.C., L.F.P., F.J.S., S.K., A.M.P. and B.L. performed analysis and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
N.-C.C. is an employee of Exai Bio. L.F.P. received financial funds from Genentech. L.F.P. received travel funds to speak at events hosted by ONT. F.J.S. received research support from Genetech, Illumina, Pacbio and ONT. S.K. has received travel funds to speak at events hosted by Oxford Nanopore Technologies for ethics conflicts. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jan Korbel, Erik Garrison and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Regions unique to T2T-CHM13 and variant calls within.
Regions unique to T2T-CHM13 compared to GRCh38 (blue) and high-quality HG002 variant calls from DeepVariant in these regions (red).
Extended Data Fig. 2 Mapping accuracy comparison using simulated data.
Mapping accuracy using simulated reads that carry GRCh38-based HG001 genotypes.
Extended Data Fig. 3 Peak memory usage comparison.
Peak memory usage of levioSAM2 and direct-to-GRC pipelines using a real 30 × WGS dataset from HG002. The alignment steps used BWA-MEM. The lift-over tasks (‘CHM13-to-GRCh38’ and ‘CHM13-to-GRCh37’) excluded the cost of the initial mapping step to T2T-CHM13 (‘CHM13’).
Extended Data Fig. 4 Small-variant calling performance using DeepVariant.
Small variant calling performance in difficult regions using DeepVariant. A. Small variant calling accuracy in major difficult genomic regions for HG002. B. GIAB stratified regions with top small variant calling error reduction densities by levioSAM2.
Extended Data Fig. 5 A disagreed SV call between the GIAB Tier 1 SV callset and personalized assemblies.
IGV visualization near chr5:21,543,010 for the HG002 PacBio-HiFi dataset. The reads were grouped using the allele at chr5:21,543,010. A 174-bp DEL was called when using direct-to-GRCh37, matching the GIAB Tier 1 SV callset. However, personalized whole-genome assemblies showed mappings of non-GRCh37 haplotypes in this region (the ‘2’ alignment in ‘HG002 Hap1’ and ‘HG002 Hap2’ tracks), suggesting collapsed mapping. The CHM13-to-GRCh37 mappings showed better concordance with the personalized HG002 assemblies.
Extended Data Fig. 6 An example of improved mapping using ONT data.
IGV visualization near chr7:125,400,000 (located in the KMT2C gene) for the HG002 ONT dataset. Four FP SV calls were made when aligning reads directly to GRCh38 because of large-scale mapping collapse. The levioSAM2 workflow (‘CHM13-to-GRCh38’) generated improved alignments and did not result in the FP SV calls.
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2, Figs. 1–9 and Tables 1–19.
Source data
Source Data Fig. 2
Variant calling summary reports generated using hap.py for HG001, HG002 and HG005 Illumina data.
Source Data Fig. 3
Stratified variant calling (GATK) summary generated using hap.py for HG002 Illumina data.
Source Data Fig. 4
Variant calling summary reports of HG002 PacBio-HiFi data.
Source Data Fig. 6
Computational efficiency reports.
Source Data Extended Data Fig. 4
Stratified variant calling (DeepVariant) summary generated using hap.py for HG002 Illumina data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, NC., Paulin, L.F., Sedlazeck, F.J. et al. Improved sequence mapping using a complete reference genome and lift-over. Nat Methods 21, 41–49 (2024). https://doi.org/10.1038/s41592-023-02069-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-023-02069-6
This article is cited by
-
Measuring, visualizing, and diagnosing reference bias with biastools
Genome Biology (2024)
-
Rapid genomic sequencing for genetic disease diagnosis and therapy in intensive care units: a review
npj Genomic Medicine (2024)