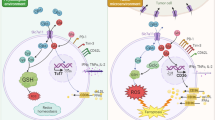
Abstract
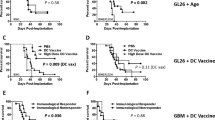
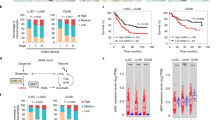
The oncometabolite (R)-2-hydroxyglutarate (R-2-HG) produced by isocitrate dehydrogenase (IDH) mutations promotes gliomagenesis via DNA and histone methylation. Here, we identify an additional activity of R-2-HG: tumor cell–derived R-2-HG is taken up by T cells where it induces a perturbation of nuclear factor of activated T cells transcriptional activity and polyamine biosynthesis, resulting in suppression of T cell activity. IDH1-mutant gliomas display reduced T cell abundance and altered calcium signaling. Antitumor immunity to experimental syngeneic IDH1-mutant tumors induced by IDH1-specific vaccine or checkpoint inhibition is improved by inhibition of the neomorphic enzymatic function of mutant IDH1. These data attribute a novel, non-tumor cell-autonomous role to an oncometabolite in shaping the tumor immune microenvironment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yang, M., Soga, T. & Pollard, P. J. Oncometabolites: Linking altered metabolism with cancer. J. Clin. Invest. 123, 3652–3658 (2013).
Erez, A. & DeBerardinis, R. J. Metabolic dysregulation in monogenic disorders and cancer—finding method in madness. Nat. Rev. Cancer 15, 440–448 (2015).
Pollard, P. J. et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 14, 2231–2239 (2005).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Mullen, A. R. & DeBerardinis, R. J. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol. Metab. 23, 552–559 (2012).
Bardella, C. et al. Expression of Idh1R132H in the murine subventricular zone stem cell niche recapitulates features of early gliomagenesis. Cancer Cell 30, 578–594 (2016).
Kaelin, W. G.Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb. Symp. Quant. Biol. 76, 335–345 (2011).
Waitkus, M. S., Diplas, B. H. & Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol. 18, 16–26 (2016).
Leonardi, R., Subramanian, C., Jackowski, S. & Rock, C. O. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J. Biol. Chem. 287, 14615–14620 (2012).
Grassian, A. R. et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 74, 3317–3331 (2014).
Sasaki, M. et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 26, 2038–2049 (2012).
Tateishi, K. et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 28, 773–784 (2015).
Reitman, Z. J. et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl Acad. Sci. USA 108, 3270–3275 (2011).
Esmaeili, M. et al. IDH1 R132H mutation generates a distinct phospholipid metabolite profile in glioma. Cancer Res. 74, 4898–4907 (2014).
Chan, S. M. et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 21, 178–184 (2015).
Fu, X. et al. 2-Hydroxyglutarate inhibits ATP synthase and mTOR signaling. Cell Metab. 22, 508–515 (2015).
Su, R. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172, 90–105 (2018).
Fathi, A. T. et al. Elevation of urinary 2-hydroxyglutarate in IDH-mutant glioma. Oncologist 21, 214–219 (2016).
Schumacher, T. et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 512, 324–327 (2014).
Rohle, D. et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 (2013).
Wang, F. et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013).
Pusch, S. et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 133, 629–644 (2017).
Losman, J. A. & Kaelin, W. G.Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Muhlhausen, C. et al. Membrane translocation of glutaric acid and its derivatives. J. Inherit. Metab. Dis. 31, 188–193 (2008).
Pajor, A. M. & Randolph, K. M. Inhibition of the Na+/dicarboxylate cotransporter by anthranilic acid derivatives. Mol. Pharmacol. 72, 1330–1336 (2007).
Kolker, S. et al. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur. J. Neurosci. 16, 21–28 (2002).
Latini, A. et al. Mitochondrial energy metabolism is markedly impaired by D-2-hydroxyglutaric acid in rat tissues. Mol. Genet. Metab. 86, 188–199 (2005).
Schumacher, T., Bunse, L., Wick, W. & Platten, M. Mutant IDH1: an immunotherapeutic target in tumors. Oncoimmunology 3, e974392 (2014).
Brown, C. E. et al. Bioactivity and safety of IL13Rα2-redirected chimeric antigen receptor CD8+T cells in patients with recurrent glioblastoma. Clin. Cancer Res. 21, 4062–4072 (2015).
Duncan, C. G. et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 22, 2339–2355 (2012).
Wick, W. et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J. Clin. Oncol. 27, 5874–5880 (2009).
Wick, W. et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro Oncol. 18, 1529–1537 (2016).
Amankulor, N. M. et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 31, 774–786 (2017).
Kohanbash, G. et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J. Clin. Invest. 127, 1425–1437 (2017).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Martinez, G. J. et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 42, 265–278 (2015).
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T Cell responses. Cell 162, 1217–1228 (2015).
Ledderose, C. et al. Mitochondria are gate-keepers of T cell function by producing the ATP that drives purinergic signaling. J. Biol. Chem. 289, 25936–25945 (2014).
Blagih, J. et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015).
Chan, A. Y. et al. Resveratrol inhibits cardiac hypertrophy via AMP-activated protein kinase and Akt. J. Biol. Chem. 283, 24194–24201 (2008).
Passariello, C. L. et al. Evidence that AMP-activated protein kinase can negatively modulate ornithine decarboxylase activity in cardiac myoblasts. Biochim. Biophys. Acta 1823, 800–807 (2012).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Ray, R. M., Bavaria, M. & Johnson, L. R. Interaction of polyamines and mTOR signaling in the synthesis of antizyme (AZ). Cell Signal. 27, 1850–1859 (2015).
Bunse, L. et al. Proximity ligation assay evaluates IDH1R132H presentation in gliomas. J. Clin. Invest. 125, 593–606 (2015).
Pellegatta, S. et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol. Commun. 3, 4 (2015).
Reardon, D. A. et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol. Res. 4, 124–135 (2016).
Chen, J. Y. et al. The oncometabolite R-2-hydroxyglutarate activates NF-κB-dependent tumor-promoting stromal niche for acute myeloid leukemia cells. Sci. Rep. 6, 32428 (2016).
Losman, J. A. et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 (2013).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8(+) T-lymphocyte fate. Nature 540, 236–241 (2016).
Olivera-Bravo, S. et al. Striatal neuronal death mediated by astrocytes from the Gcdh−/− mouse model of glutaric acidemia type I. Hum. Mol. Genet. 24, 4504–4515 (2015).
Karlstaedt, A. et al. Oncometabolite d-2-hydroxyglutarate impairs α-ketoglutarate dehydrogenase and contractile function in rodent heart. Proc. Natl Acad. Sci. USA 113, 10436–10441 (2016).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Harrington, L. E., Janowski, K. M., Oliver, J. R., Zajac, A. J. & Weaver, C. T. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452, 356–360 (2008).
Campos, B. et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin. Cancer Res. 16, 2715–2728 (2010).
Kumai, T. et al. Optimization of peptide vaccines to induce robust antitumor CD4 T-cell responses. Cancer Immunol. Res. 5, 72–83 (2017).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Buescher, J. M., Moco, S., Sauer, U. & Zamboni, N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 82, 4403–4412 (2010).
Balss, J. et al. Enzymatic assay for quantitative analysis of (D)-2-hydroxyglutarate. Acta Neuropathol. 124, 883–891 (2012).
Barber, T. W., Brockway, J. A. & Higgins, L. S. The density of tissues in and about the head. Acta Neurol. Scand. 46, 85–92 (1970).
Sahm, F. et al. Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol. 22, 26–31 (2012).
Burstenbinder, K., Rzewuski, G., Wirtz, M., Hell, R. & Sauter, M. The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 49, 238–249 (2007).
Shi, W., Oshlack, A. & Smyth, G. K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204 (2010).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462 (2016).
Hummel, M., Meister, R. & Mansmann, U. GlobalANCOVA: exploration and assessment of gene group effects. Bioinformatics 24, 78–85 (2008).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Sahm, F. et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 18, 682–694 (2017).
Acknowledgements
We acknowledge the support of the DKFZ Light Microscopy Facility, the microarray unit of the DKFZ Genomics and Proteomics Core Facility, the Transgenic Service of the Center for Preclinical Research, DKFZ, and the DKFZ–Bayer Alliance. We thank the Metabolomics Core Technology Platform of the Excellence Cluster CellNetworks for support with UPLC-based metabolite quantification. The results published here are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/. A2DR1 mice were provided by Institute Pasteur. We thank S. Kircher for flow cytometric analyses; H.Y. Ngyuen, L. Dörner, K. Rauschenbach, and M. Fischer for technical support; and J. Jung for graphics design. This work was supported by the epigenetics@dkfz program to L.B., the DKFZ-MOST program (project number 2526) and the Helmholtz Program Immunology and Inflammation, the Dr. Rolf M. Schwiete Foundation and the German Research Foundation (DFG) (FOR2289: PL315/3-1), the Sonderförderlinie ‘Neuroinflammation’ of the Ministry of Science of Baden Württemberg and the German Ministry of Education and Science (National Center for Tumor Diseases Heidelberg NCT 3.0 program ‘Precision immunotherapy of brain tumors’ and the DKTK program) to M.Pl. the Joint Funding Program MGH-Heidelberg Alliance in Neuro-Oncology to M.Pl. and M.S., the Wilhelm Sander Foundation (2012.118.1) and the German Cancer Aid (70112399) to M.Pl. and A.v.D., the German Cancer Aid (110624) to W.W., the ZUK 49/2 from the DFG to G.P., FOR2289 to B.A.N. and D.A., SFB894 to B.A.N., and the German Cancer Aid to S.T.. L.B. and M.F. are members of the MD/PhD program at Heidelberg University. L.B. was funded by Heidelberg Medical Faculty. T.B., K.S., J.K.S., and M.Ki. are supported by the Helmholtz International Graduate School, T.B. is supported by the Medical Faculty and University Hospital Mannheim. F.S. is supported by a postdoctoral fellowship of the University Hospital Heidelberg. B.W. is supported by the Faculty of Medicine of the Technical University of Munich (KKF grant). E.G. is supported by a Marie-Curie fellowship.
Author information
Authors and Affiliations
Contributions
L.B., S.P., and T.B. designed and performed experiments, analyzed data, and wrote the paper. F.S. and A.v.D. provided glioma tissue, determined IDH1 status, provided tissue stainings, and performed 850 k methylation arrays. K.S., M.F., M.Ki., A.v.L., and S.K.-B. performed in vitro experiments. J.K.S., M.Kr., and I.O. performed in vivo experiments. D.A. and B.A.N. performed calcium and respiration measurements. E.G., M.B., and R.H. performed genetic modification of cell lines. K.D. analyzed primary human tissue. C.N., M.L.S., S.U., and K.B. were involved in TIL processing. T.K. and D.S. analyzed RNA-seq data. A.S.B. and M.Pr. provided glioma tissue, determined IDH1 status, and provided immunohistochemistry stainings. K.M., M.S., D.Z., B.N., and M.D. performed metabolomics and interpreted data. B.W. performed statistical and TCGA analyses. M.O.B. performed magnetic resonance imaging. R.A.-A. and S.T. performed epigenetic profiling. J.M. and A.H. performed RNA-seq. G.P. performed adenosine phosphate and polyamine measurements. M.W. provided glioma tissues and was involved in data interpretation. M.N.-O., N.T., M.C.B., P.N.H., M.R., D.P.C., K.H.P., and D.H. provided glioma tissue. A.B. performed KEGG pathway analyses. J.E. and J.O. performed R-2-HG measurements. C.H.-M. provided the primary glioma cell line. S.K. and H.H.-S. interpreted data and provided BAY1436032. W.W. was involved in study design and data interpretation. M.Pl. conceptualized the study, interpreted data, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
M.Pl., W.W., and T.B. are inventors on a patent application entitled ‘Means and methods for treating or diagnosing IDH1R132H mutant-positive cancers’ (WO 2013/102641 A1, PCT/EP2013/050048). S.P. and A.v.D. are eligible to royalties as co-inventors of BAY 1436032 and are patent holders of ‘Means and methods for the determination of (D)-2-hydroxyglutarate (D2HG)’ (WO2013127997A1). This patent is under the administrative supervision of the DKFZ technology transfer office. K.M., M.S., D.Z., B.N., and M.D. are full-time employees of Agios. S.K. and H.H.S. are full-time employees of Bayer. The other authors declare no conflict of interest. Requests for materials should be addressed to m.platten@dkfz.de.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–5
Rights and permissions
About this article
Cite this article
Bunse, L., Pusch, S., Bunse, T. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med 24, 1192–1203 (2018). https://doi.org/10.1038/s41591-018-0095-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0095-6
This article is cited by
-
Cancer immunometabolism: advent, challenges, and perspective
Molecular Cancer (2024)
-
Revealing the role of SPP1+ macrophages in glioma prognosis and therapeutic targeting by investigating tumor-associated macrophage landscape in grade 2 and 3 gliomas
Cell & Bioscience (2024)
-
Metabolic reprogramming in the tumor microenvironment of liver cancer
Journal of Hematology & Oncology (2024)
-
Metabolic waypoints during T cell differentiation
Nature Immunology (2024)
-
Metabolic engineering for optimized CAR-T cell therapy
Nature Metabolism (2024)