Abstract
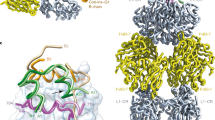
Cone snail venoms contain a wide variety of bioactive peptides, including insulin-like molecules with distinct structural features, binding modes and biochemical properties. Here, we report an active humanized cone snail venom insulin with an elongated A chain and a truncated B chain, and use cryo-electron microscopy (cryo-EM) and protein engineering to elucidate its interactions with the human insulin receptor (IR) ectodomain. We reveal how an extended A chain can compensate for deletion of B-chain residues, which are essential for activity of human insulin but also compromise therapeutic utility by delaying dissolution from the site of subcutaneous injection. This finding suggests approaches to developing improved therapeutic insulins. Curiously, the receptor displays a continuum of conformations from the symmetric state to a highly asymmetric low-abundance structure that displays coordination of a single humanized venom insulin using elements from both of the previously characterized site 1 and site 2 interactions.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-sequencing data have been deposited into the GenBank Nucleotide Database (accession numbers MW091321, MW091322, MW091323 and MW091324). Coordinates of the refined atomic models have been deposited in the Protein Data Bank (‘head’: 7MQO; ‘whole’: 7MQR; ‘asymmetric’: 7MQS). The associated cryo-EM maps have been deposited in the Electron Microscopy Data Bank (‘head’: EMD-23949; ‘whole’: EMD-23950; ‘asymmetric’: EMD-23951). Raw cryo-EM movies are available on the Electron Microscopy Public Image Archive (accession code: EMPIAR-10736). Other data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Shabanpoor, F., Separovic, F. & Wade, J. D. The human insulin superfamily of polypeptide hormones. Vitam. Horm. 80, 1–31 (2009).
Heinemann, L. Variability of insulin absorption and insulin action. Diabetes Technol. Ther. 4, 673–682 (2004).
Gradel, A. K. J. et al. Factors affecting the absorption of subcutaneously administered insulin: effect on variability. J. Diabetes Res. 2018, 1205121 (2018).
Menting, J. G. et al. How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 (2013).
Carpenter, F. H. Relationship of structure to biological activity of insulin as revealed by degradative studies. Am. J. Med. 40, 750–758 (1966).
Cruz, L. J., Gray, W. R., Yoshikami, D. & Olivera, B. M. Conus venoms—a rich source of neuroactive peptides. J. Toxicol. Toxin Rev. 4, 107–132 (1985).
Olivera, B. M. Conus venom peptide: reflections from the biology of clades and species. Annu. Rev. Ecol. Syst. 33, 25–47 (2002).
Safavi-Hemami, H. et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl Acad. Sci. USA 112, 1743–1748 (2015).
Xiong, X. et al. A structurally minimized yet fully active insulin based on cone-snail venom insulin principles. Nat. Struct. Mol. Biol. 27, 615–624 (2020).
Ahorukomeye, P. et al. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. eLife 8, e41574 (2019).
Menting, J. G. et al. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat. Struct. Mol. Biol. 23, 916–920 (2016).
Safavi-Hemami, H. et al. Venom insulins of cone snails diversify rapidly and track prey taxa. Mol. Biol. Evol. 33, 2924–2934 (2016).
Ul-Hasan, S. et al. Characterization of the peptidylglycine α-amidating monooxygenase (PAM) from the venom ducts of neogastropods, Conus bullatus and Conus geographus. Toxicon 74, 215–224 (2013).
Gutmann, T. et al. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell Biol. 219, e201907210 (2020).
Scapin, G. et al. Structure of the insulin receptor–insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 (2018).
Uchikawa, E., Choi, E., Shang, G., Yu, H. & Bai, X.-C. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor–ligand complex. eLife 8, e48630 (2019).
Weis, F. et al. The signalling conformation of the insulin receptor ectodomain. Nat. Commun. 9, 4420 (2018).
Menting, J. G. et al. Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl Acad. Sci. USA 111, E3395–E3404 (2014).
Pandyarajan, V. et al. Aromatic anchor at an invariant hormone–receptor interface. J. Biol. Chem. 289, 34709–34727 (2014).
Žáková, L. et al. Structural integrity of the B24 site in human insulin is important for hormone functionality. J. Biol. Chem. 288, 10230–10240 (2013).
Hansen, B. F., Kurtzhals, P., Jensen, A. B., Dejgaard, A. & Russell-Jones, D. Insulin X10 revisited: a super-mitogenic insulin analogue. Diabetologia 54, 2226–2231 (2011).
Schreiber, G. & Fersht, A. R. Energetics of protein–protein interactions: analysis of the Barnase–Barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248, 478–486 (1995).
Hoyne, P. A. et al. High affinity insulin binding by soluble insulin receptor extracellular domain fused to a leucine zipper. FEBS Lett. 479, 15–18 (2000).
Whittaker, J., Garcia, P., Yu, G. Q. & Mynarcik, D. C. Transmembrane domain interactions are necessary for negative cooperativity of the insulin receptor. Mol. Endocrinol. 8, 1521–1527 (1994).
Lou, M. et al. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl Acad. Sci. USA 103, 12429–12434 (2006).
Croll, T. I. et al. Higher-resolution structure of the human insulin receptor ectodomain: multi-modal inclusion of the insert domain. Structure 24, 469–476 (2016).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Brems, D. N. et al. Altering the association properties of insulin by amino acid replacement. Protein Eng. 5, 527–533 (1992).
Gammeltoft, S. et al. Insulin aspart: a novel rapid-acting human insulin analogue. Expert Opin. Investig. Drugs 8, 1431–1442 (1999).
Rakatzi, I. et al. A novel insulin analog with unique properties: LysB3, GluB29 insulin induces prominent activation of insulin receptor substrate 2, but marginal phosphorylation of insulin receptor substrate 1. Diabetes 52, 2227–2238 (2003).
Lawrence, M. C. Understanding insulin and its receptor from their three-dimensional structures. Mol. Metab. 52, 101255 (2021).
Jiráček, J. & Žáková, L. Structural perspectives of insulin receptor isoform-selective insulin analogs. Front. Endocrinol. 8, 167 (2017).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Asarnow, D., Palovcak, E. & Cheng, Y. UCSF pyem v0.5. Zenodo https://doi.org/10.5281/zenodo.3576630 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Denley, A. et al. Structural determinants for high-affinity binding of insulin-like growth factor II to insulin receptor (IR)-A, the exon 11 minus isoform of the IR. Mol. Endocrinol. 18, 2502–2512 (2004).
Menting, J. G., Ward, C. W., Margetts, M. B. & Lawrence, M. C. A thermodynamic study of ligand binding to the first three domains of the human insulin receptor: relationship between the receptor α-chain C-terminal peptide and the site 1 insulin mimetic peptides. Biochemistry 48, 5492–5500 (2009).
Lata, S., Reichel, A., Brock, R., Tampe, R. & Piehler, J. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J. Am. Chem. Soc. 127, 10205–10215 (2005).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2016).
Wickham, H., François, R., Henry, L. & Müller, K. dplyr: A Grammar of Data Manipulation (R Foundation for Statistical Computing, 2021).
Auguie, B. & Antonov, A. gridExtra: Miscellaneous Functions for “Grid” Graphics (R Foundation for Statistical Computing, 2017).
R Core Team. RStudio: Integrated Development for R (R Foundation for Statistical Computing, 2020).
Rajapaksha, H. & Forbes, B. E. Ligand-binding affinity at the insulin receptor isoform-A and subsequent IR-A tyrosine phosphorylation kinetics are important determinants of mitogenic biological outcomes. Front. Endocrinol. 6, 107 (2015).
Salic, A. & Mitchison, T. J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA 105, 2415–2420 (2008).
Gauguin, L. et al. Structural basis for the lower affinity of the insulin-like growth factors for the insulin receptor. J. Biol. Chem. 283, 2604–2613 (2008).
Disotuar, M. M. et al. Facile synthesis of insulin fusion derivatives through sortase A ligation. Acta Pharm. Sin. B 11, 2719–2725 (2020).
Hayes, D. B., Laue, T. & Philo, J. SEDNTERP http://www.jphilo.mailway.com/sednterp.htm (1995).
Schuck, P., Perugini, M. A., Gonzales, N. R., Howlett, G. J. & Schubert, D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82, 1096–1111 (2002).
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015).
Acknowledgements
We thank B.M. Olivera for cone snail collection and identification and insightful discussions, P. Shen for advice with structure determination and editing, D. Timm for EM screening and data collection at the University of Utah Electron Microscopy Core Laboratory and Paula Flórez Salcedo for the illustration of the Conus kinoshitai shell. The support and resources from the Center for High Performance Computing and the High Throughput Genomics Core Facility at the University of Utah are gratefully acknowledged. Financial support was provided by the National Institutes of Health NIDDK (DK120430 to D.H.-C.C., DK127268 to C.P.H. and DK118082 to S.J.F.), NIGMS (GM125001 to D.H.-C.C.), Juvenile Diabetes Research Foundation (5-CDA-2018-572-A-N to D.H.-C.C. and 1-INO-2017-441-A-N to H.S.-H.), German Federal Ministry of Education and Research (BMBF) grant to the German Center for Diabetes Research (DZD e.V. to Ü.C.), Deutsche Forschungsgemeinschaft (DFG 251981924–TRR 83 to Ü.C. and DFG 347368302 to Ü.C. and T.G.) and the Australian National Health and Medical Research Council (APP1143546 to M.C.L. and B.E.F.). Support of M.C.L.’s research is also made possible at WEHI through Victorian State Government Operational Infrastructure Support and the Australian NHMRC Independent Research Institutes Infrastructure Support Scheme. H.S.-H. acknowledges fellowship support from the Villum Foundation (19063) and the Carlsberg Foundation (CF19-0445).
Author information
Authors and Affiliations
Contributions
X.X. designed, synthesized and purified insulin analogs. A.B. prepared cryo-EM samples, processed cryo-EM data and modeled atomic coordinates with input from I.B.S. A.B. performed analytical ultracentrifugation with D.E. J.H.K., Y.W.Z. and X.X. performed pAkt-based activity assays. J.G.M. and M.C.L. performed the ITC experiments. H.L.S. screened cryo-EM samples and collected data sets. T.G., G.O.A. and Ü.C. expressed and purified the IR ectodomain and performed nanoDSF experiments and western blots to assess signal transduction in Hep-G2 cells. C.D., A.M. and B.E.F. performed signal transduction western blots and DNA synthesis assays in L6 myoblasts. R.A. and S.J.F. performed in vivo glucose response assays. X.X., A.B., H.S.-H., C.P.H. and D.H.-C.C. interpreted data, generated figures and wrote the manuscript with significant input from M.C.L., Ü.C., I.B.S. and T.G. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A patent application related to this work was filed by the University of Utah.
Peer review
Peer review information
Nature Chemical Biology thanks Jiří Jiráček and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
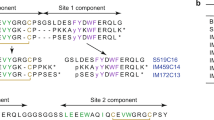
Extended Data Fig. 1 Precursor sequence alignment of venom insulins identified in this study.
Canonical arrangement of preproinsulins with N-terminal signal sequences (purple) followed by the B chain (blue), C-peptide region (black) and A chain (green). The signal sequence, C peptide(s), and additional black-colored residues are predicted to be cleaved during post-translational processing.
Extended Data Fig. 2 AKT phosphorylation activity of Vh-Ins-HTLQ and related analogs.
NIH 3T3 cells overexpressing IR-B were stimulated with insulin analogs and pAkt was quantified using a homogeneous time-resolved fluorescence assay. Error bars (s.e.m. of 4 biological replicates) are shown when larger than the symbols. Two substitutions on the B chain, GluB10 and LeuB20, were found to increase the relative activity of Vh-Ins-HTLQ. These substitutions were subsequently included in later stages of design of Vh-Ins molecules.
Extended Data Fig. 3 Vh-Ins-HSLQ at site 2.
Density is shown around Vh-Ins-HSLQ. Vh-Ins-HSLQ green, with Vh-Ins mutated residues relative to native human insulin shown in orange. Receptor FnIII-1 domain, purple. The only Vh-specific residue that approaches receptor at site 2 is GluB10, which has poor density. Nearby receptor side chains lack density but are shown explicitly for illustrative purposes.
Extended Data Fig. 4 Activity of Vh-Ins analogs with single-residue substitutions in the extended A-chain residues (A21-A24).
NIH 3T3 cells overexpressing IR-B were stimulated with insulin analogs and pAkt was quantified using a homogeneous time-resolved fluorescence assay. Error bars (s.e.m. of 4 biological replicates) are shown when larger than the symbols.
Extended Data Fig. 5 Vh-Ins-HALQ binding to IR and IGF-1R ectodomains.
a, NanoDSF monitoring of intrinsic protein fluorescence to determine the thermal conformational stability of IR-ECD (top) or IGF1R-ECD (bottom) in the presence of respective ligands in four-times molar excess. Apo-IR-ECD displays two detectable unfolding transitions Tmlow and Tmhigh at 59.2 °C and 63.2 °C, respectively (Supplementary Table 5). The presence of Vh-Ins-HALQ leads to a decrease in Tmlow to 56.3 °C indicating conformational changes induced by ligand binding similarly to insulin (Tmlow = 54.3 °C). Apo-IGF1R-ECD displays a single transition temperature Tmhigh, while binding to hIGF-I leads to an additional melting transition at 57.4 °C. No significant changes in unfolding transitions were observed for IR-ECD in the presence of hIGF-I or for IGF1R-ECD in the presence of Vh-Ins-HALQ or hIns as compared to the respective ligand-free ectodomains. b, MST with IR-ECD (left) and IGF1R-ECD (right) to determine dissociation constants of binding to respective ligands (Supplementary Table 6; n = 3, error bars show standard deviations).
Extended Data Fig. 6 Comparison of the two receptor protomers in the asymmetric conformation against previously reported structures.
Left, Vh-Ins:IR asymmetric state apolike protomer (blue) vs apo IR (PDBs 4ZXB). Right, the second Vh-Ins:IR protomer (pink) vs insulin-bound receptor (6PXV) following alignment on L1, CR, L2, and FnIII-1 domains.
Extended Data Fig. 7 Vh-Ins-HALQ signal transduction in Hep-G2 cells.
Signal transduction in Hep-G2 hepatoblastoma cells induced by Vh-Ins-HALQ and hIns at 10 or 50 nM was assessed by Western blot and densitometry (4-6 biological replicates for each condition). Phosphorylation-specific antibodies were used to detect phosphorylated IR (Y1150/115), IGF1R (Y980), AKT1 (S473), and MEK-1/2 (S217/221). Relative intensities of specific protein bands were normalized to the GAPDH loading control and then to the respective signal after 5 min of insulin treatment.
Extended Data Fig. 8 SV-AUC c(s) analysis of insulin analogs.
a, DOI (des octapeptide insulin) and hIns (human insulin) controls at a concentration of 100 µg/ml and 775 µg/ml respectively in phosphate buffer (137 mM NaCl, 2.7 mM KCl, 5.3 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). b, Vh-Ins-HSLQ and Lispro both at 500 µg/ml in sterile insulin diluent (16 mg/ml glycerol, 1.6 mg/ml m-cresol, 0.65 mg/ml phenol, 3.8 mg/ml Na2HPO4, pH 7.4). c-d, Data fit and residuals for DOI, hIns, Vh-Ins-HSLQ and lispro, respectively. For clarity, some scans are omitted from the figures shown but all scans were used for the c(s) analysis. The interval between the scans shown in each panel is ~9 minutes.
Extended Data Fig. 9 SV-AUC of DOI and hIns in sterile diluent.
a, c(s) analysis of DOI and hIns in sterile diluent. For reference the Vh-Ins-HSLQ trace from Fig. S11 is shown. b-c, data fit and residuals for DOI and hIns in sterile diluent, respectively. hIns shows increasing concentrations at higher radii in early scans, indicative of aggregation during the experiment.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Note and Tables 1–6.
Supplementary Video 1
A series of volumes derived from 3D variability analysis show a range of flexible motion in one of the two receptor protomers. Models were built into the conformational extremes and used to create a series of molecular motions that were fit into the experimental density. In the asymmetric extreme, three Vh-Ins molecules are apparent in the complex, including one in a composite site 1/site 2 position. As the receptor moves toward the symmetric state, L1 and CR move away from their position near the composite site, and the separate density for both site 1- and site 2-bound Vh-Ins becomes apparent.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Xiong, X., Blakely, A., Kim, J.H. et al. Symmetric and asymmetric receptor conformation continuum induced by a new insulin. Nat Chem Biol 18, 511–519 (2022). https://doi.org/10.1038/s41589-022-00981-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-00981-0
This article is cited by
-
Structural conservation of insulin/IGF signalling axis at the insulin receptors level in Drosophila and humans
Nature Communications (2023)
-
A stepwise activation model for the insulin receptor
Experimental & Molecular Medicine (2023)
-
Recombinant Insulin-Like Growth Factor 1 Dimers: Receptor Binding Affinities and Activation Abilities
International Journal of Peptide Research and Therapeutics (2023)
-
Unconventional insulins from predators and pathogens
Nature Chemical Biology (2022)
-
Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nature Communications (2022)