Abstract
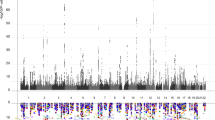
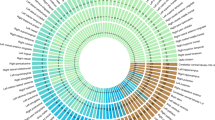
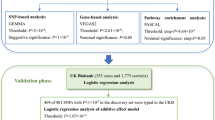
The hippocampus is critical for memory and cognition and neuropsychiatric disorders, and its subfields differ in architecture and function. Genome-wide association studies on hippocampal and subfield volumes are mainly conducted in European populations; however, other ancestral populations are under-represented. Here we conduct cross-ancestry genome-wide association meta-analyses in 65,791 individuals for hippocampal volume and 38,977 for subfield volumes, including 7,009 individuals of East Asian ancestry. We identify 339 variant–trait associations at P < 1.13 × 10−9 for 44 hippocampal traits, including 23 new associations. Common genetic variants have similar effects on hippocampal traits across ancestries, although ancestry-specific associations exist. Cross-ancestry analysis improves the fine-mapping precision and the prediction performance of polygenic scores in under-represented populations. These genetic variants are enriched for Wnt signaling and neuron differentiation and affect cognition, emotion and neuropsychiatric disorders. These findings may provide insight into the genetic architectures of hippocampal and subfield volumes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The GWAS summary statistics used in this work from following two publicly available datasets: the UKBB study (https://open.win.ox.ac.uk/ukbiobank/big40/) and the ENIGMA study (https://enigma.ini.usc.edu/research/download-enigma-gwas-results/data-agreement-for-hippocampal-volume-gwas-download/). All GWAS summary statistics from EAS-specific and cross-ancestry meta-analyses of the 44 hippocampal and subfield volumes are publicly available on http://chimgen.tmu.edu.cn/en/index.php?c=article&id=2359, http://www.mulinlab.org/Hipp_img_TA_meta/ and the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/home) with the accession numbers: GCST90267894–GCST90267981. The PGSs are publicly available on the PGS Catalog (https://www.pgscatalog.org/) with the ID of PGP000464 and score IDs of PGS003591–PGS003722.
Code availability
We made use of publicly available software and tools. All codes used to generate results reported in this paper are publicly available at GitHub (https://github.com/Nana-Liu-genetics/Protocols) and Zenodo (https://doi.org/10.5281/zenodo.7847774)94.
References
Scoville, W. B. & Milner, B. Loss of recent memory after bilateral hippocampal lesions. J. Neurol. Neurosurg. Psychiatry 20, 11–21 (1957).
Morris, R. G., Garrud, P., Rawlins, J. N. & O’Keefe, J. Place navigation impaired in rats with hippocampal lesions. Nature 297, 681–683 (1982).
Lisman, J. et al. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nat. Neurosci. 20, 1434–1447 (2017).
Kim, J. J. & Diamond, D. M. The stressed hippocampus, synaptic plasticity and lost memories. Nat. Rev. Neurosci. 3, 453–462 (2002).
Moreno-Jiménez, E. P. et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560 (2019).
Li, J. Q. et al. Risk factors for predicting progression from mild cognitive impairment to Alzheimer’s disease: a systematic review and meta-analysis of cohort studies. J. Neurol. Neurosurg. Psychiatry 87, 476–484 (2016).
Calabresi, P., Castrioto, A., Di Filippo, M. & Picconi, B. New experimental and clinical links between the hippocampus and the dopaminergic system in Parkinson’s disease. Lancet Neurol. 12, 811–821 (2013).
Mattai, A. et al. Hippocampal volume development in healthy siblings of childhood-onset schizophrenia patients. Am. J. Psychiatry 168, 427–435 (2011).
Treadway, M. T. et al. Illness progression, recent stress, and morphometry of hippocampal subfields and medial prefrontal cortex in major depression. Biol. Psychiatry 77, 285–294 (2015).
Gonçalves, J. T., Schafer, S. T. & Gage, F. H. Adult neurogenesis in the hippocampus: from stem. Cells Behav. Cell 167, 897–914 (2016).
Kühn, S. et al. Plasticity of hippocampal subfield volume cornu ammonis 2+3 over the course of withdrawal in patients with alcohol dependence. JAMA Psychiatry 71, 806–811 (2014).
Pitman, R. K. et al. Biological studies of post-traumatic stress disorder. Nat. Rev. Neurosci. 13, 769–787 (2012).
Fanselow, M. S. & Dong, H. W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65, 7–19 (2010).
Iglesias, J. E. et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137 (2015).
Ho, N. F. et al. Progression from selective to general involvement of hippocampal subfields in schizophrenia. Mol. Psychiatry 22, 142–152 (2017).
Small, S. A., Schobel, S. A., Buxton, R. B., Witter, M. P. & Barnes, C. A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 12, 585–601 (2011).
Hibar, D. P. et al. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Bis, J. C. et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat. Genet. 44, 545–551 (2012).
Stein, J. L. et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 44, 552–561 (2012).
Hibar, D. P. et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 8, 13624 (2017).
Elliott, L. T. et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018).
Zhao, B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 51, 1637–1644 (2019).
Smith, S. M. et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 24, 737–745 (2021).
van der Meer, D. et al. Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Mol. Psychiatry 25, 3053–3065 (2020).
Zahodne, L. B. et al. Structural MRI predictors of late-life cognition differ across African Americans, Hispanics, and Whites. Curr. Alzheimer Res. 12, 632–639 (2015).
Li, M. et al. Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol. Psychiatry 19, 452–461 (2014).
Li, M. et al. Failure of replicating the association between hippocampal volume and 3 single-nucleotide polymorphisms identified from the European genome-wide association study in Asian populations. Neurobiol. Aging 35, 2883 (2014).
Peterson, R. E. et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179, 589–603 (2019).
Li, Y. R. & Keating, B. J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 6, 91 (2014).
Chen, M. H. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213 (2020).
Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504 (2018).
Chen, J. et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 53, 840–860 (2021).
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019).
Xu, Q. et al. CHIMGEN: a Chinese imaging genetics cohort to enhance cross-ethnic and cross-geographic brain research. Mol. Psychiatry 25, 517–529 (2020).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Thompson, P. M. et al. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry 10, 100 (2020).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Del Villar, K. & Miller, C. A. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc. Natl Acad. Sci. USA 101, 4210–4215 (2004).
Mann, F., Chauvet, S. & Rougon, G. Semaphorins in development and adult brain: implication for neurological diseases. Prog. Neurobiol. 82, 57–79 (2007).
Kichaev, G. & Pasaniuc, B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97, 260–271 (2015).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Benner, C. et al. Prospects of fine-mapping trait-associated genomic regions by using summary statistics from genome-wide association studies. Am. J. Hum. Genet. 101, 539–551 (2017).
Kanai, M. et al. Meta-analysis fine-mapping is often miscalibrated at single-variant resolution. Cell Genom. 2, 100210 (2022).
Towers, E. et al. The proapoptotic dp5 gene is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons. Nucleic Acids Res. 37, 3044–3060 (2009).
Consortium, G. T. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Litterman, N. et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 9, e1001060 (2011).
Nishino, J., Kim, I., Chada, K. & Morrison, S. J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239 (2008).
Patwari, P. et al. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 284, 24996–25003 (2009).
Graham, S. E. et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 600, 675–679 (2021).
Duncan, L. et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 10, 3328 (2019).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Liu, Y. et al. APOE genotype and neuroimaging markers of Alzheimer’s disease: systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 86, 127–134 (2015).
Belloy, M. E., Napolioni, V. & Greicius, M. D. A quarter century of APOE and Alzheimer’s disease: progress to date and the path forward. Neuron 101, 820–838 (2019).
Matissek, S. J. & Elsawa, S. F. GLI3: a mediator of genetic diseases, development and cancer. Cell Commun. Signal. 18, 54 (2020).
Wilson, P. M., Fryer, R. H., Fang, Y. & Hatten, M. E. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J. Neurosci. 30, 8529–8540 (2010).
Lionel, A. C. et al. Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum. Mol. Genet. 23, 2752–2768 (2014).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
O’Brien, H. E. et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 19, 194 (2018).
O’Brien, H. et al. 55Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Eur. Neuropsychopharmacol. 29, S1098–S1099 (2019).
Takemoto, T. et al. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394–398 (2011).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–w205 (2019).
Jacobs, S. et al. Mice with targeted Slc4a10 gene disruption have small brain ventricles and show reduced neuronal excitability. Proc. Natl Acad. Sci. USA 105, 311–316 (2008).
Sinning, A., Liebmann, L. & Hübner, C. A. Disruption of Slc4a10 augments neuronal excitability and modulates synaptic short-term plasticity. Front. Cell. Neurosci. 9, 223 (2015).
Sun, C. et al. Nonenzymatic function of DPP4 in diabetes-associated mitochondrial dysfunction and cognitive impairment. Alzheimers Dement. 18, 966–987 (2021).
Hussaini, S. M. et al. Wnt signaling in neuropsychiatric disorders: ties with adult hippocampal neurogenesis and behavior. Neurosci. Biobehav. Rev. 47, 369–383 (2014).
Miller, K. L. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A. L. et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83, 132–135 (2008).
Anderson, C. A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Wu, D. et al. Large-scale whole-genome sequencing of three diverse Asian populations in Singapore. Cell 179, 736–749 (2019).
König, I. R., Loley, C., Erdmann, J. & Ziegler, A. How to include chromosome X in your genome-wide association study. Genet. Epidemiol. 38, 97–103 (2014).
Fortin, J. P. et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161, 149–170 (2017).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011).
Buniello, A. et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Luo, Y. et al. Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. Hum. Mol. Genet. 30, 1521–1534 (2021).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience 8, giz082 (2019).
Amendola, L. M. et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 25, 305–15 (2015).
Hinrichs, A. S. The UCSC genome browser database: update 2006. Nucleic Acids Res. 34, D590–D598 (2006).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Liu, N. Nana-Liu-genetics/Protocols: Protocols_Nana. Zenodo https://doi.org/10.5281/zenodo.7847774 (2023).
Acknowledgements
We are grateful to the UKBB and ENIGMA for providing GWAS summary statistics of hippocampal and subfield volumes. This work was supported by the National Natural Science Foundation of China (82030053 and 81425013 to C.Y.).
Author information
Authors and Affiliations
Consortia
Contributions
C.Y. and N.L. designed the study and wrote the paper. All authors critically reviewed the paper. N.L., L.Z., T.T., J.C., B.Z., S.Q., M.J.L., K.X., X-N.Z., M.W., Z. Ye, W.Q., F.L., M.L., Q.X., J.F., J.X. and C.Y. were the principal investigators. L.Z., T.T., J.C., B.Z., S.Q., Z.G., G.C., Q.Z., W. Liao, Y.Y., H.Z., B.G, X.X., T.H., Z. Yao, W.Z., P.Z., W. Li, D.S., C.W., S.L., Z. Yan, F.C., J.L., J.Z., D.W., W.S., Y.M., J.X., J-H.G., X.Z., K.X., X-N.Z., M.W., Z. Ye and C.Y. acquired the data. C.Y., Z. Ye and M.W. supervised this work. N.L. analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Jason Stein and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10.
Supplementary Tables
Supplementary Tables 1–21.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, N., Zhang, L., Tian, T. et al. Cross-ancestry genome-wide association meta-analyses of hippocampal and subfield volumes. Nat Genet 55, 1126–1137 (2023). https://doi.org/10.1038/s41588-023-01425-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-023-01425-8