Abstract
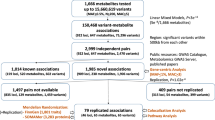
In cross-platform analyses of 174 metabolites, we identify 499 associations (P < 4.9 × 10−10) characterized by pleiotropy, allelic heterogeneity, large and nonlinear effects and enrichment for nonsynonymous variation. We identify a signal at GLP2R (p.Asp470Asn) shared among higher citrulline levels, body mass index, fasting glucose-dependent insulinotropic peptide and type 2 diabetes, with β-arrestin signaling as the underlying mechanism. Genetically higher serine levels are shown to reduce the likelihood (by 95%) and predict development of macular telangiectasia type 2, a rare degenerative retinal disease. Integration of genomic and small molecule data across platforms enables the discovery of regulators of human metabolism and translation into clinical insights.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All genome-wide summary statistics are made available through an interactive web server (https://omicscience.org/apps/crossplatform/) and were further uploaded to the GWAS catalog (https://www.ebi.ac.uk/gwas/, accession numbers GCST90010722–GCST90010862).
Code availability
Source code was deposited in the following repository: https://github.com/MRC-Epid/crossplatform_mGWAS.
References
Wishart, D. S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 99, 1819–1875 (2019).
Shin, S.-Y. Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Long, T. et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49, 568–578 (2017).
Draisma, H. H. M. et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 6, 7208 (2015).
Kettunen, J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 7, 11122 (2016).
Illig, T. et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42, 137–141 (2010).
Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 (2011).
Rhee, E. P. P. et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 18, 130–143 (2013).
Gallois, A. et al. A comprehensive study of metabolite genetics reveals strong pleiotropy and heterogeneity across time and context. Nat. Commun. 10, 4788 (2019).
Rhee, E. P. et al. An exome array study of the plasma metabolome. Nat. Commun. 7, 12360 (2016).
Astle, W. J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429 (2016).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Learn, D. B., Fried, V. A. & Thomas, E. L. Taurine and hypotaurine content of human leukocytes. J. Leukoc. Biol. 48, 174–182 (1990).
Yet, I. et al. Genetic influences on metabolite levels: a comparison across metabolomic platforms. PLoS ONE 11, e0153672 (2016).
Foley, C. N. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Preprint at bioRiv https://doi.org/10.1101/592238 (2019).
Pedersen, C. B. et al. The ACADS gene variation spectrum in 114 patients with short-chain acyl-CoA dehydrogenase (SCAD) deficiency is dominated by missense variations leading to protein misfolding at the cellular level. Hum. Genet. 124, 43–56 (2008).
Lahiri, S. et al. Kinetic characterization of mammalian ceramide synthases: determination of Km values towards sphinganine. FEBS Lett. 581, 5289–5294 (2007).
Horowitz, B. et al. Asparagine synthetase activity of mouse leukemias. Science 160, 533–535 (1968).
Babu, E. et al. Identification of a novel system l amino acid transporter structurally distinct from heterodimeric amino acid transporters. J. Biol. Chem. 278, 43838–43845 (2003).
Scott, R. A. et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 66, 2888–2902 (2017).
Wheeler, E. et al. Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med. 14, e1002383 (2017).
Prokopenko, I. et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10, e1004235 (2014).
Manning, A. K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 (2012).
Almgren, P. et al. Genetic determinants of circulating GIP and GLP-1 concentrations. JCI Insight 2, e93306 (2017).
Fragkos, K. C. & Forbes, A. Citrulline as a marker of intestinal function and absorption in clinical settings: a systematic review and meta-analysis. United European Gastroenterol. J. 6, 181–191 (2018).
Tseng, C. C. & Zhang, X. Y. The cysteine of the cytoplasmic tail of glucose-dependent insulinotropic peptide receptor mediates its chronic desensitization and down-regulation. Mol. Cell. Endocrinol. 139, 179–186 (1998).
Estall, J. L., Koehler, J. A., Yusta, B. & Drucker, D. J. The glucagon-like peptide-2 receptor C terminus modulates β-arrestin-2 association but is dispensable for ligand-induced desensitization, endocytosis, and G-protein-dependent effector activation. J. Biol. Chem. 280, 22124–22134 (2005).
Scerri, T. S. et al. Genome-wide analyses identify common variants associated with macular telangiectasia type 2. Nat. Genet. 49, 559–567 (2017).
Gantner, M. L. et al. Serine and lipid metabolism in macular disease and peripheral neuropathy. N. Engl. J. Med. 381, 1422–1433 (2019).
Garrod, A. E. The incidence of alkaptonuria: a study in chemical individuality. Lancet 160, 1616–1620 (1902).
Rath, A. et al. Representation of rare diseases in health information systems: the orphanet approach to serve a wide range of end users. Hum. Mutat. 33, 803–808 (2012).
Stübiger, G. et al. Targeted profiling of atherogenic phospholipids in human plasma and lipoproteins of hyperlipidemic patients using MALDI-QIT-TOF-MS/MS. Atherosclerosis 224, 177–186 (2012).
van der Graaf, A., Kastelein, J. J. P. & Wiegman, A. Heterozygous familial hypercholesterolaemia in childhood: cardiovascular risk prevention. J. Inherit. Metab. Dis. 32, 699 (2009).
Lindsay, T. et al. Descriptive epidemiology of physical activity energy expenditure in UK adults (the Fenland study). Int. J. Behav. Nutr. Phys. Act. 16, 126 (2019).
Day, N. et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br. J. Cancer 80, 95–103 (1999).
Moore, C. et al. The INTERVAL trial to determine whether intervals between blood donations can be safely and acceptably decreased to optimise blood supply: study protocol for a randomised controlled trial. Trials 15, 363 (2014).
Soininen, P. et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 134, 1781–1785 (2009).
Wittemans, L. B. L. et al. Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat. Commun. 10, 1060 (2019).
Lotta, L. A. et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 13, e1002179 (2016).
Di Angelantonio, E. et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45,000 donors. Lancet 390, 2360–2371 (2017).
Li, J. & Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Stacey, D. et al. ProGeM: a framework for the prioritization of candidate causal genes at molecular quantitative trait loci. Nucleic Acids Res. 47, e3 (2019).
Wishart, D. S. et al. HMDB 4.0: the Human Metabolome Database for 2018. Nucleic Acids Res. 46, D608–D617 (2018).
Bateman, A. et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Burgess, S. & Thompson, S. G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 181, 251–260 (2015).
Harrow, J. et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012).
Lee, J. J. Y., Wasserman, W. W., Hoffmann, G. F., Van Karnebeek, C. D. M. & Blau, N. Knowledge base and mini-expert platform for the diagnosis of inborn errors of metabolism. Genet. Med. 20, 151–158 (2018).
Köhler, S. et al. The human phenotype ontology in 2017. Nucleic Acids Res. 45, D865–D876 (2017).
Wu, P. et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform. 7, e14325 (2019).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Acknowledgements
M.P. was supported by a fellowship from the German Research Foundation (DFG PI 1446/2-1). C.O.-W. was funded by an early career fellowship at Homerton College, University of Cambridge. L.B.L.W. acknowledges funding from the Wellcome Trust (WT083442AIA). J.L.G. was supported by grants from the Medical Research Council (MC_UP_A090_1006, MC_PC_13030, MR/P011705/1 and MR/P01836X/1). Work in the Reimann and Gribble laboratories was supported by the Wellcome Trust (106262/Z/14/Z and 106263/Z/14/Z), the UK Medical Research Council (MRC_MC_UU_12012/3) and PhD funding for E.K.B. from MedImmune/AstraZeneca. P.S. is supported by a Rutherford Fund Fellowship from the Medical Research Council (MR/S003746/1). A.M.W. is supported by a BHF-Turing Cardiovascular Data Science Award and by the EC-Innovative Medicines Initiative (BigData@Heart). J.R. is supported by the German Federal Ministry of Education and Research (BMBF) within the framework of e:Med research and funding concept (grant no. 01ZX1912D). J.D. is funded by the National Institute for Health Research (NIHR; Senior Investigator Award) (*). The EPIC-Norfolk study (https://doi.org/10.22025/2019.10.105.00004) received funding from the Medical Research Council (MR/N003284/1 and MC-UU_12015/1) and Cancer Research UK (C864/A14136). The genetic work in the EPIC-Norfolk study was funded by the Medical Research Council (MC_PC_13048). Metabolite measurements in the EPIC-Norfolk study were supported by the MRC Cambridge Initiative in Metabolic Science (MR/L00002/1) and the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement no. 115372. The Fenland study is supported by the UK Medical Research Council (MC_UU_12015/1 and MC_PC_13046). Nightingale Health NMR assays were funded by the European Commission Framework Programme 7 (HEALTH-F2-2012-279233). Metabolon Metabolomics assays, the academic coordinating center, DNA extraction and genotyping for INTERVAL were supported by core funding from the NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014-10024), the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (SP/09/002, RG/13/13/30194 and RG/18/13/33946), the NIHR (Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust) and the NIHR BioResource (http://bioresource.nihr.ac.uk) (*). This work was supported by Health Data Research UK, which is funded by the UK Medical Research Council, the Engineering and Physical Sciences Research Council, the Economic and Social Research Council, the Department of Health and Social Care (England), the Chief Scientist Office of the Scottish Government Health and Social Care Directorates, the Health and Social Care Research and Development Division (Welsh Government), the Public Health Agency (Northern Ireland), the British Heart Foundation and Wellcome. We are grateful to all the participants who have been part of the project and to the many members of the study teams at the University of Cambridge who enabled this research. *The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. UK Biobank: this research was conducted using the UK Biobank resource under application no. 44448.
Author information
Authors and Affiliations
Consortia
Contributions
L.A.L. and C. Langenberg designed the study. L.A.L., M.P. and C. Langenberg drafted the manuscript. L.A.L., M.P., I.D.S., L.B.L.W., C. Li, R.B., C.O.-W, V.P.W.A., J.L., E.W., E.P., P.S., S.B., V.Z. and E.S. analyzed the data. J.R. and G.K. designed and implemented the web server. K.-T.K. and N.J.W. are principal investigators of the EPIC-Norfolk cohort. G.A.M. advised on metabolite mapping across platforms. A.K. and F.I. provided metabolite measurements and quality control in the Fenland study. E.K.B., F.M.G. and F.R. performed all experimental work on GLP2R. M.B. contributed data for MacTel. E.F. performed knowledge-based annotation of genes to variants. J.D. and A.S.B. were responsible for the INTERVAL study. All authors contributed to the interpretation of results and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.S.B. received grants from AstraZeneca, Biogen, Bioverativ, Merck, Novartis and Sanofi. J.D. sits on the International Cardiovascular and Metabolic Advisory Board for Novartis (since 2010), the Steering Committee of UK Biobank (since 2011), is a member of the MRC International Advisory Group, London (since 2013), the MRC High Throughput Science ’Omics Panel, London (since 2013) and serves on the Scientific Advisory Committee for Sanofi (since 2013), the International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis and the AstraZeneca Genomics Advisory Board (since 2018). E.F. is an employee and stock holder of Pfizer. L.A.L. is presently an employee and shareholder of Regeneron Pharmaceuticals Inc. The remaining authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1–5
Supplementary Tables
Supplementary Tables 1–6
Supplementary Data 1
Sample size for each metabolite.
Supplementary Data 2
Results for MR analysis on MacTel.
Rights and permissions
About this article
Cite this article
Lotta, L.A., Pietzner, M., Stewart, I.D. et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet 53, 54–64 (2021). https://doi.org/10.1038/s41588-020-00751-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-00751-5
This article is cited by
-
Multi-omic prediction of incident type 2 diabetes
Diabetologia (2024)
-
Genome-wide characterization of circulating metabolic biomarkers
Nature (2024)
-
Impact of the gut microbiota and associated metabolites on cardiometabolic traits, chronic diseases and human longevity: a Mendelian randomization study
Journal of Translational Medicine (2023)
-
Genetically predicted plasma levels of amino acids and metabolic dysfunction-associated fatty liver disease risk: a Mendelian randomization study
BMC Medicine (2023)
-
Assessing the causal effect of genetically predicted metabolites and metabolic pathways on stroke
Journal of Translational Medicine (2023)