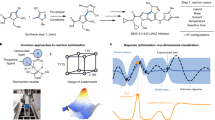
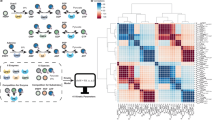
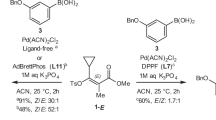
Abstract
Reaction conditions that are generally applicable to a wide variety of substrates are highly desired, especially in the pharmaceutical and chemical industries1,2,3,4,5,6. Although many approaches are available to evaluate the general applicability of developed conditions, a universal approach to efficiently discover these conditions during optimizations is rare. Here we report the design, implementation and application of reinforcement learning bandit optimization models7,8,9,10 to identify generally applicable conditions by efficient condition sampling and evaluation of experimental feedback. Performance benchmarking on existing datasets statistically showed high accuracies for identifying general conditions, with up to 31% improvement over baselines that mimic state-of-the-art optimization approaches. A palladium-catalysed imidazole C–H arylation reaction, an aniline amide coupling reaction and a phenol alkylation reaction were investigated experimentally to evaluate use cases and functionalities of the bandit optimization model in practice. In all three cases, the reaction conditions that were most generally applicable yet not well studied for the respective reaction were identified after surveying less than 15% of the expert-designed reaction space.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All reaction datasets evaluated in simulation studies and the two newly collected reaction datasets (the palladium-catalysed C–H arylation reaction and the amide coupling reaction) are available at GitHub (https://github.com/doyle-lab-ucla/bandit-optimization). Raw data logs from simulation studies with both synthetic data and chemistry reaction data are available at Zenodo (https://doi.org/10.5281/zenodo.8170874).
Code availability
All source codes for implemented optimization algorithms and models, simulation methods for synthetic data and chemistry reaction dataset and analysis functions for data logs and optimization results are available at GitHub (https://github.com/doyle-lab-ucla/bandit-optimization). The current release of the software is also available at Zenodo (https://doi.org/10.5281/zenodo.8181283).
References
Wagen, C. C., McMinn, S. E., Kwan, E. E. & Jacobsen, E. N. Screening for generality in asymmetric catalysis. Nature 610, 680–686 (2022).
Rein, J. et al. Generality-oriented optimization of enantioselective aminoxyl radical catalysis. Science 380, 706–712 (2023).
Betinol, I. O., Lai, J., Thakur, S. & Reid, J. P. A data-driven workflow for assigning and predicting generality in asymmetric catalysis. J. Am. Chem. Soc. 145, 12870–12883 (2023).
Kim, H. et al. A multi-substrate screening approach for the identification of a broadly applicable Diels–Alder catalyst. Nat. Commun. 10, 770 (2019).
Angello, N. H. et al. Closed-loop optimization of general reaction conditions for heteroaryl Suzuki-Miyaura coupling. Science 378, 399–405 (2022).
Rinehart, N. I. et al. A machine-learning tool to predict substrate-adaptive conditions for Pd-catalyzed C–N couplings. Science 381, 965–972 (2023).
Lattimore, T. & Szepesvári, C. Bandit Algorithms (Cambridge Univ. Press, 2020).
Sutton, R. S. & Barto, A. G. Reinforcement Learning: An Introduction 2nd edn (Bradford Books, 2018).
Slivkins, A. Introduction to multi-armed bandits. Preprint at arxiv.org/abs/1904.07272v7 (2019).
White, J. M. Bandit Algorithms for Website Optimization: Developing, Deploying, and Debugging (O’Reilly Media, 2013).
Ruiz-Castillo, P. & Buchwald, S. L. Applications of palladium-catalyzed C–N cross-coupling reactions. Chem. Rev. 116, 12564–12649 (2016).
Ogba, O. M., Warner, N. C., O’Leary, D. J. & Grubbs, R. H. Recent advances in ruthenium-based olefin metathesis. Chem. Soc. Rev. 47, 4510–4544 (2018).
Kolb, H. C., VanNieuwenhze, M. S. & Sharpless, K. B. Catalytic asymmetric dihydroxylation. Chem. Rev. 94, 2483–2547 (1994).
Chatterjee, S., Guidi, M., Seeberger, P. H. & Gilmore, K. Automated radial synthesis of organic molecules. Nature 579, 379–384 (2020).
Echtermeyer, A., Amar, Y., Zakrzewski, J. & Lapkin, A. Self-optimisation and model-based design of experiments for developing a C–H activation flow process. Beilstein J. Org. Chem. 13, 150–163 (2017).
Coley, C. W., Abolhasani, M., Lin, H. & Jensen, K. F. Material‐efficient microfluidic platform for exploratory studies of visible‐light photoredox catalysis. Angew. Chem. Int. Ed. 56, 9847–9850 (2017).
Granda, J. M., Donina, L., Dragone, V., Long, D.-L. & Cronin, L. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature 559, 377–381 (2018).
Hsieh, H.-W., Coley, C. W., Baumgartner, L. M., Jensen, K. F. & Robinson, R. I. Photoredox iridium-nickel dual catalyzed decarboxylative arylation cross-coupling: from batch to continuous flow via self-optimizing segmented flow reactor. Org. Process Res. Dev. 22, 542–550 (2018).
Schweidtmann, A. M. et al. Machine learning meets continuous flow chemistry: automated optimization towards the Pareto front of multiple objectives. Chem. Eng. J. 352, 277–282 (2018).
Burger, B. et al. A mobile robotic chemist. Nature 583, 237–241 (2020).
Häse, F., Aldeghi, M., Hickman, R. J., Roch, L. M. & Aspuru-Guzik, A. Gryffin: an algorithm for Bayesian optimization of categorical variables informed by expert knowledge. Appl. Phys. Rev. 8, 031406 (2021).
Taylor, C. J. et al. Accelerated chemical reaction optimization using multi-task learning. ACS Cent. Sci. 9, 957–968 (2023).
Zhou, Z., Li, X. & Zare, R. N. Optimizing chemical reactions with deep reinforcement learning. ACS Cent. Sci. 3, 1337–1344 (2017).
Torres, J. A. G. et al. A multi-objective active learning platform and web app for reaction optimization. J. Am. Chem. Soc. 144, 19999–20007 (2022).
Shields, B. J. et al. Bayesian reaction optimization as a tool for chemical synthesis. Nature 590, 89–96 (2021).
Häse, F., Roch, L. M., Kreisbeck, C. & Aspuru-Guzik, A. Phoenics: a Bayesian optimizer for chemistry. ACS Cent. Sci. 4, 1134–1145 (2018).
Clayton, A. D. et al. Algorithms for the self-optimisation of chemical reactions. React. Chem. Eng. 4, 1545–1554 (2019).
Reker, D., Hoyt, E. A., Bernardes, G. J. L. & Rodrigues, T. Adaptive optimization of chemical reactions with minimal experimental information. Cell Rep. Phys. Sci. 1, 100247 (2020).
Shim, E. et al. Predicting reaction conditions from limited data through active transfer learning. Chem. Sci. 13, 6655–6668 (2022).
Gao, H. et al. Using machine learning to predict suitable conditions for organic reactions. ACS Cent. Sci. 4, 1465–1476 (2018).
Kozlowski, M. C. On the topic of substrate scope. Org. Lett. 24, 7247–7249 (2022).
Gensch, T. & Glorius, F. The straight dope on the scope of chemical reactions. Science 352, 294–295 (2016).
Dreher, S. D. Catalysis in medicinal chemistry. React. Chem. Eng. 4, 1530–1535 (2019).
Kariofillis, S. K. et al. Using data science to guide aryl bromide substrate scope analysis in a Ni/photoredox-catalyzed cross-coupling with acetals as alcohol-derived radical sources. J. Am. Chem. Soc. 144, 1045–1055 (2022).
Dreher, S. D. & Krska, S. W. Chemistry informer libraries: conception, early experience, and role in the future of cheminformatics. Acc. Chem. Res. 54, 1586–1596 (2021).
Collins, K. D. & Glorius, F. A robustness screen for the rapid assessment of chemical reactions. Nat. Chem. 5, 597–601 (2013).
Kullmer, C. N. P. et al. Accelerating reaction generality and mechanistic insight through additive mapping. Science 376, 532–539 (2022).
Taylor, C. J. et al. A brief introduction to chemical reaction optimization. Chem. Rev. 123, 3089–3126 (2023).
Svensson, H. G., Bjerrum, E. J., Tyrchan, C., Engkvist, O. & Chehreghani, M. H. Autonomous drug design with multi-armed bandits. In 2022 IEEE International Conference on Big Data 5584–5592 (IEEE, 2022).
Romeo Atance, S., Viguera Diez, J., Engkvist, O., Olsson, S. & Mercado, R. De novo drug design using reinforcement learning with graph-based deep generative models. J. Chem. Inf. Model. 62, 4863–4872 (2022).
Xu, Z., Shim, E., Tewari, A. & Zimmerman, P. Adaptive sampling for discovery. In Proc. Advances in Neural Information Processing System Vol. 35, 1114–1126 (NeurIPS, 2022).
Kaufmann, E., Cappe, O. & Garivier, A. On Bayesian upper confidence bounds for bandit problems. In Proc. Machine Learning Research Vol. 22, 592–600 (PMLR, 2012).
Auer, P., Cesa-Bianchi, N. & Fischer, P. Finite-time analysis of the multiarmed bandit problem. Mach. Learn. 47, 235–256 (2002).
Snoek, J. et al. Scalable Bayesian optimization using deep neural networks. In Proc. Machine Learning Research Vol. 27, 2171–2180 (PMLR, 2015).
Stevens, J. M. et al. Advancing base metal catalysis through data science: insight and predictive models for Ni-catalyzed borylation through supervised machine learning. Organometallics 41, 1847–1864 (2022).
Nielsen, M. K., Ahneman, D. T., Riera, O. & Doyle, A. G. Deoxyfluorination with sulfonyl fluorides: navigating reaction space with machine learning. J. Am. Chem. Soc. 140, 5004–5008 (2018).
Lin, S. et al. Mapping the dark space of chemical reactions with extended nanomole synthesis and MALDI-TOF MS. Science 361, eaar6236 (2018).
Ahneman, D. T., Estrada, J. G., Lin, S., Dreher, S. D. & Doyle, A. G. Predicting reaction performance in C–N cross-coupling using machine learning. Science 360, 186–190 (2018).
Brown, D. G. & Boström, J. Analysis of past and present synthetic methodologies on medicinal chemistry: where have all the new reactions gone? J. Med. Chem. 59, 4443–4458 (2016).
El-Faham, A. & Albericio, F. Peptide coupling reagents, more than a letter soup. Chem. Rev. 111, 6557–6602 (2011).
Dombrowski, A. W., Aguirre, A. L., Shrestha, A., Sarris, K. A. & Wang, Y. The chosen few: parallel library reaction methodologies for drug discovery. J. Org. Chem. 87, 1880–1897 (2022).
Matheron, G. Principles of geostatistics. Econ. Geol. 58, 1246–1266 (1963).
Zimmerman, D., Pavlik, C., Ruggles, A. & Armstrong, M. P. An experimental comparison of ordinary and universal kriging and inverse distance weighting. Math. Geol. 31, 375–390 (1999).
Magano, J. Large-scale amidations in process chemistry: practical considerations for reagent selection and reaction execution. Org. Process Res. Dev. 26, 1562–1689 (2022).
Beutner, G. L. et al. TCFH–NMI: direct access to N-acyl imidazoliums for challenging amide bond formations. Org. Lett. 20, 4218–4222 (2018).
Stevens, J. M. et al. Leveraging high-throughput experimentation to drive pharmaceutical route invention: a four-step commercial synthesis of branebrutinib (BMS-986195). Org. Process Res. Dev. 26, 1174–1183 (2022).
Sperry, J. B. et al. Thermal stability assessment of peptide coupling reagents commonly used in pharmaceutical manufacturing. Org. Process Res. Dev. 22, 1262–1275 (2018).
Zheng, B. et al. Preparation of the HIV attachment inhibitor BMS-663068. Part 6. Friedel–Crafts acylation/hydrolysis and amidation. Org. Process Res. Dev. 21, 1145–1155 (2017).
Krishnan, K. K., Ujwaldev, S. M., Sindhu, K. S. & Anilkumar, G. Recent advances in the transition metal catalyzed etherification reactions. Tetrahedron 72, 7393–7407 (2016).
Fuhrmann, E. & Talbiersky, J. Synthesis of alkyl aryl ethers by catalytic Williamson ether synthesis with weak alkylation agents. Org. Process Res. Dev. 9, 206–211 (2005).
Swamy, K. C. K., Kumar, N. N. B., Balaraman, E. & Kumar, K. V. P. P. Mitsunobu and related reactions: advances and applications. Chem. Rev. 109, 2551–2651 (2009).
Acknowledgements
The financial support for this study was provided by BMS, the Princeton Catalysis Initiative, the NSF under the CCI Center for Computer Assisted Synthesis (CHE-2202693) and the Dreyfus Program for Machine Learning in the Chemical Sciences and Engineering. J.Y.W. acknowledges support from the BMS Graduate Fellowship in Synthetic Organic Chemistry. S.K.K. acknowledges support from the NSF Graduate Research Fellowship Program under grant no. DGE-1656466. M.P. acknowledges support from the NIH F32 Ruth L. Kirschstein NRSA Fellowship (1F32GM129910-01A1). We thank J. Raab, M. Ruos and S. Gandhi for reviewing the Supplementary Information.
Author information
Authors and Affiliations
Contributions
J.Y.W. and A.G.D. designed the overall research project. J.Y.W. designed and implemented optimization models and algorithms with inputs from J.M.S., J.L., J.E.T., B.J.S. and A.G.D.; J.M.S., B.J.S., J.L., J.E.T., J.Y.W. and A.G.D. designed and planned reaction scopes for the C–H arylation reaction, the amide coupling reaction and the phenol alkylation reaction. J.M.S., S.K.K., M.-J.T., D.L.G., M.P., D.N.P., B.H., D.D., S.D., A.F., G.G.Z., S.M. and J.P. carried out high-throughput experiments and authentic product synthesis for the three reactions. J.Y.W. wrote the paper with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Jolene Reid and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Testing the bandit algorithms on a previously published C–N cross-coupling reaction dataset.
a, General reaction scheme of the C–N cross-coupling reaction and reactivity heatmap grouped by base and ligand, with average yields for each base/ligand combination shown in white text. Structures for all substrates and conditions in the scope are included in the Supplementary Information. b, Top three most general base–ligand conditions for the dataset. c, Average accuracies of identifying top-3 conditions with various algorithms across 500 simulations with random starts. Exploration refers to the uniform exploration required by some algorithms, during which each condition is sequentially selected once. Different implementations of TS and Bayes UCB algorithms were used and differentiated by implementation 1 and 2 for simplicity. This plot is reproduced in Fig. S83, with the details of the algorithms included in the legend. TS: Thompson Sampling; UCB: upper confidence bound. d, Real-time optimization progress for simulation 0 (the first simulation) of a Bayes UCB (implementation 2) algorithm at n = 12, 30, 60, 99. Squares with different colors represent all reactions that have been suggested and evaluated by the algorithm at the time. The real-time empirical average for each base/ligand combination is shown in white texts.
Extended Data Fig. 2 Model architecture and workflow of bandit algorithms during reaction optimization.
The bandit algorithm suggests a condition (an arm) to evaluate first. The chemist-designed reaction scope suggests a reaction to evaluate with the selected condition. The suggested reaction is tested experimentally, and the result is used to update both the reaction scope and the bandit algorithm for the next round of proposal. Finally, a prediction model, separately trained with existing experimental results, is optionally used to propose reactions to evaluate via other mechanisms (e.g., batch proposal).
Supplementary information
Supplementary Information
Supplementary Sections 1–12, including Supplementary Text and Data, Supplementary Figs. 1–119 and Supplementary Tables 1–3 – see Contents pages for details.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J.Y., Stevens, J.M., Kariofillis, S.K. et al. Identifying general reaction conditions by bandit optimization. Nature 626, 1025–1033 (2024). https://doi.org/10.1038/s41586-024-07021-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-07021-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.