Abstract
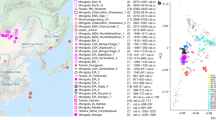
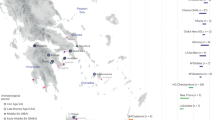
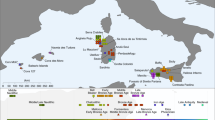
Soqotra, an island situated at the mouth of the Gulf of Aden in the northwest Indian Ocean between Africa and Arabia, is home to ~60,000 people subsisting through fishing and semi-nomadic pastoralism who speak a Modern South Arabian language. Most of what is known about Soqotri history derives from writings of foreign travellers who provided little detail about local people, and the geographic origins and genetic affinities of early Soqotri people has not yet been investigated directly. Here we report genome-wide data from 39 individuals who lived between ~650 and 1750 ce at six locations across the island and document strong genetic connections between Soqotra and the similarly isolated Hadramawt region of coastal South Arabia that likely reflects a source for the peopling of Soqotra. Medieval Soqotri can be modelled as deriving ~86% of their ancestry from a population such as that found in the Hadramawt today, with the remaining ~14% best proxied by an Iranian-related source with up to 2% ancestry from the Indian sub-continent, possibly reflecting genetic exchanges that occurred along with archaeologically documented trade from these regions. In contrast to all other genotyped populations of the Arabian Peninsula, genome-level analysis of the medieval Soqotri is consistent with no sub-Saharan African admixture dating to the Holocene. The deep ancestry of people from medieval Soqotra and the Hadramawt is also unique in deriving less from early Holocene Levantine farmers and more from groups such as Late Pleistocene hunter–gatherers from the Levant (Natufians) than other mainland Arabians. This attests to migrations by early farmers having less impact in southernmost Arabia and Soqotra and provides compelling evidence that there has not been complete population replacement between the Pleistocene and Holocene throughout the Arabian Peninsula. Medieval Soqotra harboured a small population that showed qualitatively different marriage practices from modern Soqotri, with first-cousin unions occurring significantly less frequently than today.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The aligned sequences are available through the European Nucleotide Archive under accession number PRJEB66485. Genotype data are available at https://reich.hms.harvard.edu/datasets. Any other relevant data are available from the corresponding authors upon reasonable request.
Code availability
The custom code used in this study is available at https://github.com/DReichLab/ADNA-Tools.
References
Jansen van Rensburg, J. et al. The Soqotra Heritage Project (2017–2020). Bull. Soc. Arab. Stud. 22, 26–27 (2022).
Strauch, I. Foreign Sailors on Socotra: The Inscriptions and Drawings from the Cave Hoq (Hempen, 2012).
Naumkin, V. V. A. Island of the Phoenix: An Ethnographic Study of the People of Socotra (Paul & Company Pub Consortium, 1993).
Weeks, L., Morris, M., McCall, B. & Al-Zubairy, K. A recent archaeological survey on Soqotra. Report on the preliminary expedition season, January 5th–February 2nd 2001. Arab. Archaeol. Epigr. 13, 95–125 (2002).
Casson, L. The Periplus Maris Erythraei: Text with Introduction, Translation, and Commentary (Princeton Univ. Press, 1989).
Doe, B. Socotra: Island of Tranquillity (Immel, 1992).
Morris, M., Watson, J. C. E. & Eades, D. A Comparative Cultural Glossary across the Modern South Arabian Language Family (Oxford Univ. Press, 2019).
Evers, K. Cave of revelations: Indian Ocean trade in light of the Socotran graffiti. J. Indian Ocean Archaeol. 10, 19–37 (2014).
Bent, J. T. Southern Arabia (Smith, Elder and Co., 1900).
Doe, B. Socotra; an Archaeological Reconnaissance in 1967 (Field Research Projects, 1970).
Jansen van Rensburg, J. Rock art of Soqotra, Yemen: a forgotten heritage revisited. Arts 7, 99 (2018).
Naumkin, V. V. & Sedov, A. V. Monuments of Socotra. Topoi Orient-Occident 3, 569–623 (1993).
Shinnie, P. L. Socotra. Antiquity 34, 100–110 (1960).
Pinhasi, R. et al. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS ONE 10, e0129102 (2015).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016).
Vyas, D. N., Al-Meeri, A. & Mulligan, C. J. Testing support for the northern and southern dispersal routes out of Africa: an analysis of Levantine and southern Arabian populations. Am. J. Phys. Anthropol. 164, 736–749 (2017).
Hellenthal, G. et al. A genetic atlas of human admixture history. Science 343, 747–751 (2014).
Fedele, F. G. in The Evolution of Human Populations in Arabia: Paleoenvironments, Prehistory and Genetics (eds Petraglia, M. D. & Rose, J. I.) 215–236 (Springer, 2010).
McCorriston, J., & Martin, L. in The Evolution of Human Populations in Arabia: Paleoenvironments, Prehistory and Genetics (eds Petraglia, M. D. & Rose, J. I.) 237–250 (Springer, 2010).
Uerpmann, H.-P., Potts, D., & Uerpmann, M. in The Evolution of Human Populations in Arabia: Paleoenvironments, Prehistory and Genetics (eds Petraglia, M. D. & Rose, J. I.) 205–214 (Springer, 2010).
Almarri, M. A. et al. The genomic history of the Middle East. Cell 184, 4612–4625. e4614 (2021).
Lazaridis, I. et al. The genetic history of the Southern Arc: a bridge between West Asia and Europe. Science 377, eabm4247 (2022).
Lazaridis, I. et al. Ancient DNA from Mesopotamia suggests distinct pre-pottery and pottery Neolithic migrations into Anatolia. Science 377, 982–987 (2022).
Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016).
Černý, V. et al. Internal diversification of mitochondrial haplogroup r0a reveals post-last glacial maximum demographic expansions in South Arabia. Mol. Biol. Evol. 28, 71–78 (2011).
Agranat-Tamir, L. et al. The genomic history of the bronze age southern levant. Cell 181, 1146–1157 (2020).
Harney, É. et al. Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat. Commun. 9, 1–11 (2018).
Lazaridis, I. et al. A genetic probe into the ancient and medieval history of Southern Europe and West Asia. Science 377, 940–951 (2022).
van de Loosdrecht, M. et al. Pleistocene north African genomes link near eastern and sub-Saharan African human populations. Science 360, 548–552 (2018).
Soares, P. The expansion of mtDNA haplogroup L3 within and out of Africa. Mol. Biol. Evol. 29, 915–927 (2012).
Černý, V., Čížková, M., Poloni, E. S., Al-Meeri, A. & Mulligan, C. Comprehensive view of the population history of Arabia as inferred by mtDNA variation. Am. J. Phys. Anthropol. 159, 607–616 (2016).
Narasimhan, V. M. et al. The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019).
Shinde, V. et al. An ancient Harappan genome lacks ancestry from steppe pastoralists or Iranian farmers. Cell 179, 729–735 (2019).
Derenko, M. et al. Complete mitochondrial DNA diversity in Iranians. PLoS ONE 8, e80673 (2013).
Al-Abri, A. et al. Pleistocene–Holocene boundary in southern Arabia from the perspective of human mtDNA variation. Am. J. Phys. Anthropol. 149, 21–28 (2012).
Černý, V. et al. Out of Arabia—the settlement of Island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity. Am. J. Phys. Anthropol. 138, 439–447 (2009).
Gandini, F. et al. Mapping human dispersals into the Horn of Africa from Arabian Ice Age refugia using mitogenomes. Sci. Rep. 6, 25472–25472 (2016).
Ringbauer, H., Novembre, J. & Steinrücken, M. Parental relatedness through time revealed by runs of homozygosity in ancient DNA. Nat. Commun. 12, 5425 (2021).
Ceballos, F. C., Joshi, P. K., Clark, D. W., Ramsay, M. & Wilson, J. F. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 19, 220–234 (2018).
Cadenas, A. M., Zhivotovsky, L. A., Cavalli-Sforza, L. L., Underhill, P. A. & Herrera, R. J. Y-chromosome diversity characterizes the Gulf of Oman. Eur. J. Hum. Genet. 16, 374–386 (2008).
Ariano, B. et al. Ancient maltese genomes and the genetic geography of neolithic Europe. Curr. Biol. 32, 2668–2680.e6 (2022).
Wellsted, J. R. Travels to the City of the Caliphs, Along the Shores of the Persian Gulf and the Mediterranean: Including a Voyage to the Coast of Arabia, and a Tour on the Island of Socotra (2 volumes) (Henry Colburn, 1840).
Luciani, M., Binder, M. & Alsaud, A. S. Life and living conditions in north-west Arabia during the Bronze Age: first results from the bioarchaeological work at Qurayyah. Proceedings of the Seminar Arabian Studies 48, 185–20 (2018).
Olalde, I. et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234 (2019).
Dridi, H. Indiens et proche-orientaux dans une grotte de Suquṭrā (Yemen). J. Asiatique 290, 565–610 (2002).
Dridi, H. & Gorea, M. Au IIIe siècle de notre ère: le voyage d’Agbar à Suqutra. Archeologia 396, 48–57 (2003).
Robin, C. J. & Gorea, M. Les vestiges antiques de la grotte de Hôq (Suqutra, Yémen)(note d’information). C. R. Séances Acad. Inscr. B.-Lett. 146, 409–445 (2002).
Strauch, I. & Bukharin, M. Indian inscriptions from the Cave Ḥoq on Suquṭrā. Ann. dell’Istituto Universitario Orient. Napoli 64, 121–138 (2004).
Strauch, I. in Ports of the Ancient Indian Ocean (eds. Boussac, M.-F. et al.) 79–97 (Primus Books, 2016).
Al-Mujāwir, I., & ibn Ya’qūb, Y. A Traveller in Thirteenth-Century Arabia: Ibn al-Mujāwir’s Tārīkh al-Mustabṣir (Ashgate, 2008).
Bittles, A. H., & Hamamy, H. A. in Genetic Disorders among Arab Populations (ed. Teebi, A. S.) 85–108 (Springer, 2010).
Korotayev, A. Parallel-cousin (FBD) marriage, islamization, and arabization. Ethnology 39, 395–407 (2000).
Stern, G. H. Marriage in Early Islam (Royal Asiatic Society, 1939).
Yule, H., & Cordier, H. The Travels of Marco Polo: The Complete Yule-Cordier Edition: Including the Unabridged Third Edition (1903) of Henry Yule’s Annotated Translation, as Revised by Henri Cordier, Together with Cordier’s Later Volume of Notes and Addenda (1920) (Dover Publications/Constable, 1993).
Hansen, H. B. et al. Comparing ancient DNA preservation in petrous bone and tooth cementum. PLoS ONE 12, e0170940 (2017).
Pinhasi, R., Fernandes, D. M., Sirak, K. & Cheronet, O. Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat. Protoc. 14, 1194–1205 (2019).
Rohland, N., Glocke, I., Aximu-Petri, A. & Meyer, M. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 13, 2447–2461 (2018).
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B 370, 20130624 (2015).
Fu, Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015).
Fu, Q. et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227 (2013).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015).
Behar, D. M. et al. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Fu, Q. et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559 (2013).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: analysis of next generation sequencing data. BMC Bioinform. 15, 356 (2014).
Kennett, D. J. et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 8, 14115 (2017).
Lohse, J. C., Madsen, D. B., Culleton, B. J. & Kennett, D. J. Isotope paleoecology of episodic mid-to-late Holocene bison population expansions in the Southern Plains, USA. Quat. Sci. Rev. 102, 14–26 (2014).
Van Klinken, G. J. Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J. Archaeol. Sci. 26, 687–695 (1999).
Santos, G. M., Southon, J. R., Druffel-Rodriguez, K. C., Griffin, S. & Mazon, M. Magnesium perchlorate as an alternative water trap in AMS graphite sample preparation: a report on sample preparation at KCCAMS at the University of California, Irvine. Radiocarbon 46, 165–173 (2004).
Stuiver, M. & Polach, H. A. Discussion reporting of 14C data. Radiocarbon 19, 355–363 (1977).
Bronk Ramsey, C. & Lee, S. Recent and planned developments of the program OxCal. Radiocarbon 55, 720–730 (2013).
Reimer, P. J. et al. The IntCal20 Northern Hemisphere radiocarbon age calibration curve (0–55 cal kBP). Radiocarbon 62, 725–757 (2020).
Brielle, E. S. et al. Entwined African and Asian genetic roots of medieval peoples of the Swahili coast. Nature 615, 866–873 (2023).
Fan, S. et al. African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations. Genome Biol. 20, 1–14 (2019).
Fregel, R. et al. Ancient genomes from north Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc. Natl Acad. Sci. USA 115, 6774–6779 (2018).
Fu, Q. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014).
Gallego-Llorente, M. et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820–822 (2015).
Jones, E. R. et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912 (2015).
Kılınç, G. M. et al. The demographic development of the first farmers in Anatolia. Curr. Biol. 26, 2659–2666 (2016).
Lipson, M. et al. Ancient West African foragers in the context of African population history. Nature 577, 665–670 (2020).
Lipson, M. et al. Ancient DNA and deep population structure in sub-Saharan African foragers. Nature 603, 290–296 (2022).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012).
Prendergast, M. E. et al. Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science 365, eaaw6275 (2019).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Raghavan, M. et al. Upper palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014).
Schlebusch, C. M. et al. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358, 652–655 (2017).
Schuenemann, V. J. et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat. Commun. 8, 1–11 (2017).
Sirak, K. A. et al. Social stratification without genetic differentiation at the site of Kulubnarti in Christian Period Nubia. Nat. Commun. 12, 7283 (2021).
Skoglund, P. et al. Genetic evidence for two founding populations of the Americas. Nature 525, 104–108 (2015).
Skoglund, P. et al. Reconstructing prehistoric African population structure. Cell 171, 59–71 (2017).
Biagini, S. A. et al. People from Ibiza: an unexpected isolate in the Western Mediterranean. Eur. J. Hum. Genet. 27, 941–951 (2019).
Broushaki, F. et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 353, 99–503 (2016).
Changmai, P. et al. Indian genetic heritage in Southeast Asian populations. PLoS Genet. 18, e1010036 (2022).
Flegontov, P. et al. Palaeo-Eskimo genetic ancestry and the peopling of Chukotka and North America. Nature 570, 236–240 (2019).
Jeong, C. et al. The genetic history of admixture across inner Eurasia. Nat. Ecol. Evol. 3, 966–976 (2019).
Lipson, M. et al. Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95 (2018).
Liu, D. et al. Extensive ethnolinguistic diversity in Vietnam reflects multiple sources of genetic diversity. Mol. Biol. Evol. 37, 2503–2519 (2020).
López, S. et al. Evidence of the interplay of genetics and culture in Ethiopia. Nat. Commun. 12, 3581 (2021).
Nakatsuka, N. et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 49, 1403–1407 (2017).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Pickrell, J. K. et al. The genetic prehistory of southern Africa. Nat. Commun. 3, 1143 (2012).
Qin, P. & Stoneking, M. Denisovan ancestry in east Eurasian and Native American populations. Mol. Biol. Evol. 32, 2665–2674 (2015).
Skoglund, P. et al. Genomic insights into the peopling of the Southwest Pacific. Nature 538, 510–513 (2016).
Wang, C.-C. et al. Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419 (2021).
Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Consortium, T. G. P. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Acknowledgements
We thank N. Adamski, R. Bernardos and Z. Zhang for contributions to data generation, processing and curation. We acknowledge the help and support we received from the Soqotra Heritage Project team, including A. E. A. S. Al-Rumali, M. T. H. Ali, A. S. S. A. Al-Ameri and S. M. O. Sulaiman. We thank N. Salem for translation of the manuscript title and abstract to Arabic. The work of J. Jansen van Rensburg in Soqotra was supported by the National Geographic Society (NGS-61123R-19). D. Reich is an Investigator of the Howard Hughes Medical Institute (HHMI), and the ancient DNA laboratory work and analyses were also supported by National Institutes of Health grant HG012287, by John Templeton Foundation grant 61220, by the Allen Discovery Center program, which is a Paul G. Allen Frontiers Group advised program of the Paul G. Allen Family Foundation, and by a gift from J.-F. Clin. This article is subject to HHMI’s Open Access to Publications policy. HHMI lab heads have previously granted a nonexclusive CC BY 4.0 licence to the public and a sublicensable licence to HHMI in their research articles. Pursuant to those licences, the author-accepted manuscript of this article can be made freely available under a CC BY 4.0 licence immediately upon publication.
Author information
Authors and Affiliations
Contributions
K.S., J.J.V.R. and D.R. designed the research. J.J.V.R., A.S.A.A.-O., E.M.A.S., A.M.S.H. and D.C.B. excavated and/or contributed to the acquisition and description of the osteological and archaeological data. K.S., E.B., B.C., I.L., D.R. and H.R. carried out population genetics analyses. M.M., S.M., A.M. and A.K. contributed to data generation, processing and curation. K.C., E.C., A.M.L., J.N.W. and F.Z. performed laboratory work under the supervision of N.R. K.S. and J.J.V.R wrote and revised the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Diyendo Massilani, Miranda Morris and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–10.
Supplementary Data 1–17
Supplementary Data 1–17.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sirak, K., Jansen Van Rensburg, J., Brielle, E. et al. Medieval DNA from Soqotra points to Eurasian origins of an isolated population at the crossroads of Africa and Arabia. Nat Ecol Evol 8, 817–829 (2024). https://doi.org/10.1038/s41559-024-02322-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-024-02322-x