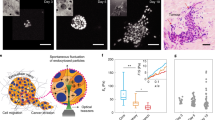
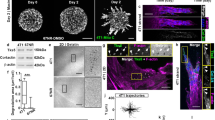
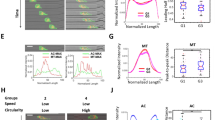
Abstract
Plasticity of cancer invasion and metastasis depends on the ability of cancer cells to switch between collective and single-cell dissemination, controlled by cadherin-mediated cell–cell junctions. In clinical samples, E-cadherin-expressing and -deficient tumours both invade collectively and metastasize equally, implicating additional mechanisms controlling cell–cell cooperation and individualization. Here, using spatially defined organotypic culture, intravital microscopy of mammary tumours in mice and in silico modelling, we identify cell density regulation by three-dimensional tissue boundaries to physically control collective movement irrespective of the composition and stability of cell–cell junctions. Deregulation of adherens junctions by downregulation of E-cadherin and p120-catenin resulted in a transition from coordinated to uncoordinated collective movement along extracellular boundaries, whereas single-cell escape depended on locally free tissue space. These results indicate that cadherins and extracellular matrix confinement cooperate to determine unjamming transitions and stepwise epithelial fluidization towards, ultimately, cell individualization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The authors declare that data supporting the findings of this study are available within the paper (and its supplementary information files), and are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
The detailed mathematical procedures, the simulation code and videos of example realizations for in silico modelling were deposited at GitHub (https://github.com/sisyga/jamminglgca). Custom MATLAB scripts for cell-nuclei tracking and cell segmentation are available at https://github.com/steffengrosser/smart-3D-registration-2019 and https://github.com/JuergenLippoldt/On-Motility-and-Form. Codes for the PIV analysis are available at https://github.com/ComplexityBiosystems/PIV-spheroids.
References
Mayor, R. & Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17, 97–109 (2016).
Friedl, P., Locker, J., Sahai, E. & Segall, J. E. Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777–783 (2012).
Gomez, G. A., McLachlan, R. W. & Yap, A. S. Productive tension: force-sensing and homeostasis of cell–cell junctions. Trends Cell Biol. 21, 499–505 (2011).
Reffay, M. et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol. 16, 217–223 (2014).
Ewald, A. J., Brenot, A., Duong, M., Chan, B. S. & Werb, Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell 14, 570–581 (2008).
Sunyer, R. et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science 353, 1157–1161 (2016).
Lehembre, F. et al. NCAM-induced focal adhesion assembly: a functional switch upon loss of E-cadherin. EMBO J. 27, 2603–2615 (2008).
Wendt, M. K., Taylor, M. A., Schiemann, B. J. & Schiemann, W. P. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol. Biol. Cell 22, 2423–2435 (2011).
Perl, A. K., Wilgenbus, P., Dahl, U., Semb, H. & Christofori, G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 392, 190–193 (1998).
Ye, X. & Weinberg, R. A. Epithelial–mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 25, 675–686 (2015).
Derksen, P. W. B. et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 (2006).
Khalil, A. A. et al. Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin. Exp. Metastasis 34, 421–429 (2017).
Kowalski, P. J., Rubin, M. A. & Kleer, C. G. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 5, R217–R222 (2003).
Narendra, S., Jenkins, S. M., Khoor, A. & Nassar, A. Clinical outcome in pleomorphic lobular carcinoma: a case-control study with comparison to classic invasive lobular carcinoma. Ann. Diagn. Pathol. 19, 64–69 (2015).
Padmanaban, V. et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019).
Beerling, E. et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 14, 2281–2288 (2016).
Fischer, K. R. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Bi, D., Yang, X., Marchetti, M. C. & Manning, M. Motility-driven glass and jamming transitions in biological tissues. Phys. Rev. X 6, 021011 (2016).
Haeger, A., Krause, M., Wolf, K. & Friedl, P. Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim. Biophys. Acta 1840, 2386–2395 (2014).
Doxzen, K. et al. Guidance of collective cell migration by substrate geometry. Integr. Biol. 5, 1026–1035 (2013).
Gaggioli, C. et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9, 1392–1400 (2007).
Weigelin, B., Bakker, G. & Friedl, P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital 1, 32–43 (2012).
van Helvert, S., Storm, C. & Friedl, P. Mechanoreciprocity in cell migration. Nat. Cell Biol. 20, 8–20 (2018).
Friedl, P. & Mayor, R. Tuning collective cell migration by cell–cell junction regulation. Cold Spring Harb. Perspect. Biol. 9, a029199 (2017).
Duclos, G. et al. Spontaneous shear flow in confined cellular nematics. Nat. Phys. 14, 728–732 (2018).
Wolf, K. et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9, 893–904 (2007).
Ilina, O. et al. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model Mech. 11, dmm034330 (2018).
Provenzano, P. P. et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6, 11 (2008).
Wolf, K. et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 (2013).
Szabo, A. et al. In vivo confinement promotes collective migration of neural crest cells. J. Cell Biol. 213, 543–555 (2016).
Balzer, E. M. et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 26, 4045–4056 (2012).
Haeger, A. et al. Collective cancer invasion forms an integrin-dependent radioresistant niche. J. Exp. Med. 217, e20181184 (2020).
Bi, D., Lopez, J. H., Schwarz, J. M. & Manning, M. L. A density-independent rigidity transition in biological tissues. Nat. Phys. 11, 1074 (2015).
Park, J.-A. et al. Unjamming and cell shape in the asthmatic airway epithelium. Nat. Mater. 14, 1040–1048 (2015).
Palamidessi, A. et al. Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma. Nat. Mater. 18, 1252–1263 (2019).
Ilina, O., Bakker, G.-J., Vasaturo, A., Hofmann, R. M. & Friedl, P. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Phys. Biol. 8, 15010 (2011).
Singhai, R. et al. E-Cadherin as a diagnostic biomarker in breast cancer. N. Am. J. Med. Sci. 3, 227–233 (2011).
Vos, C. B. et al. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br. J. Cancer 76, 1131–1133 (1997).
Conklin, M. W. et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011).
Provenzano, P. P. et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 (2006).
Elisha, Y., Kalchenko, V., Kuznetsov, Y. & Geiger, B. Dual role of E-cadherin in the regulation of invasive collective migration of mammary carcinoma cells. Sci. Rep. 8, 4986 (2018).
Cadwell, C. M., Su, W. & Kowalczyk, A. P. Cadherin tales: Regulation of cadherin function by endocytic membrane trafficking. Traffic 17, 1262–1271 (2016).
Frisch, U., Hasslacher, B. & Pomeau, Y. Lattice-gas automata for the Navier–Stokes equation. Phys. Rev. Lett. 56, 1505–1508 (1986).
Deutsch, A. & Dormann, S. Cellular Automaton Modeling of Biological Pattern Formation (Birkhäuser, 2018).
Bussemaker, H. J., Deutsch, A. & Geigant, E. Mean-field analysis of a dynamical phase transition in a cellular automaton model for collective motion. Phys. Rev. Lett. 78, 5018–5021 (1997).
Deutsch, A. A new mechanism of aggregation in a lattice-gas cellular automaton model. Math. Comput. Modell. 31, 35–40 (2000).
Duclos, G., Garcia, S., Yevick, H. G. & Silberzan, P. Perfect nematic order in confined monolayers of spindle-shaped cells. Soft Matter 10, 2346–2353 (2014).
Schotz, E.-M., Lanio, M., Talbot, J. A. & Manning, M. L. Glassy dynamics in three-dimensional embryonic tissues. J. R. Soc. Interface 10, 20130726 (2013).
Notbohm, J. et al. Cellular contraction and polarization drive collective cellular motion. Biophys. J. 110, 2729–2738 (2016).
Atia, L. et al. Geometric constraints during epithelial jamming. Nat. Phys. 14, 613–620 (2018).
Hayer, A. et al. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 18, 1311–1323 (2016).
Bazellieres, E. et al. Control of cell–cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 17, 409–420 (2015).
Angelini, T. E. et al. Glass-like dynamics of collective cell migration. Proc. Natl Acad. Sci. USA 108, 4714–4719 (2011).
Garcia, S. et al. Physics of active jamming during collective cellular motion in a monolayer. Proc. Natl. Acad. Sci. USA 112, 15314–15319 (2015).
Douezan, S. et al. Spreading dynamics and wetting transition of cellular aggregates. Proc. Natl Acad. Sci. USA 108, 7315–7320 (2011).
Truong, H. H. et al. β1 integrin inhibition elicits a prometastatic switch through the TGFβ-miR-200-ZEB network in E-cadherin-positive triple-negative breast cancer. Sci. Signal. 7, ra15 (2014).
Shamir, E. R. et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 204, 839–856 (2014).
Aiello, N. M. et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695 (2018).
Kuriyama, S. et al. In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J. Cell Biol. 206, 113–127 (2014).
Wiesner, C. et al. KIF5B and KIF3A/KIF3B kinesins drive MT1–MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood 116, 1559–1569 (2010).
Nagelkerke, A. et al. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother. Oncol. 101, 39–45 (2011).
Alieva, M., Ritsma, L., Giedt, R. J., Weissleder, R. & van Rheenen, J. Imaging windows for long-term intravital imaging: general overview and technical insights. Intravital 3, e29917 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529 (2017).
Tinevez, J.-Y. et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Thielicke, W. & Stamhuis, E. J. PIVlab—towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2, e30 (2014).
Acknowledgements
We gratefully acknowledge the Microscopical Imaging Centre core support by H. Croes and J. Fransen; M. K. te Lindert for scanning electron microscopy; B. Lemmers-Van de Weem for the excellent technical assistance in animal experiments; and L. Wolf for 3D rendering of carcinoma invasion in patient samples. We thank the Centre for Information Services and High Performance Computing at TU Dresden for providing computational infrastructure. This work was supported by Deutsche Krebshilfe (106950 and 70112014), the European Union FP7 Tissue Transmigration Training Network (T3Net 237946), the Netherlands Science Organization (NWO-VICI 918.11.626), the European Research Council (617430-DEEPINSIGHT), NIH-U54 CA210184-01, and the Cancer Genomics Center (CGC.nl) to P.F.; the German Research Foundation (Deutsche Forschungsgemeinschaft, SFB-TR 79) to A.D.; the European Social Fund (ESF), co-financed by tax funds on the basis of the budget adopted by the members of the Saxon State Parliament, to S.S.; the European Research Council (ERC-741350) to J.A.K.; AIRC under IG2018-ID 21558 (principal investigator, M. Pusch) to C.A.M.L.P.; and the Lise–Meitner fellowship M 2450-NBL by the Austrian Science Fund (FWF) to O.C.
Author information
Authors and Affiliations
Contributions
O.I.: study design, acquisition of data, analysis and interpretation of results and writing the manuscript; P.G.G.: acquisition of data, analysis and interpretation of results and writing the manuscript; S.S.: acquisition of in silico data, and analysis and interpretation of results on mathematical modelling; M.V.: acquisition of data on metastasis, and analysis and interpretation of results on metastasis; G.-J.B.: assistance with two-photon microscopy; J.S.: generation of mathematical model; P.B.: contribution of clinical tissue samples, interpretation of data and revising the manuscript; A.D.: design of mathematical model, acquisition of in silico data, analysis and interpretation of results on mathematical modelling; J.A.K., J.L. and S.G.: cell tracking and cell segmentation analysis; O.C., C.A.M.L.P. and S.Z.: particle imaging, velocimetry and data analysis; P.F.: study design, interpretation of results and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 3D organization of normal and mammary carcinoma tissue and the geometry of interstitial tissue tracks.
a, Immunofluorescence staining of E-cadherin or pan-cytokeratin (pan-CK), vimentin and collagen IV in normal ducts, IDC and ILC lesions. Arrowheads indicate single carcinoma cells. Example images represent 3 patient samples per condition. b, 3D reconstruction of z-projection (200 μm thickness) from IDC and ILC samples with carcinoma cells oriented in the niches formed by aligned collagen fibers. Example images represent 7 (IDC) and 3 (ILC) patient samples. c, 3D rendering of collective carcinoma strands and tissue tracks in IDC (left panel), and a different region with tissue tracks formed by collagen bundles (right panel). d,Tissue structure surrounding E-cadherin positive duct in normal breast tissue. Example image represents 3 normal breast tissue patient samples. e, Inter-fiber distance of tissue tracks formed by collagen bundles with stroma cells present or absent within the tracks. Data represent n = 49 and 24, 139 and 78, and 107 and 93 cell-free or cell-filled tissue microtracks pooled from 3 (normal breast tissue), 7 (tumor-free IDC tissue) and 3 (tumor-free ILC tissue) patient samples, respectively. Median (red line) and dots representing individual tissue microtracks. P values, two-tailed Mann-Whitney test. Scale bars, 50 µm. For statistical source data, see Source data for ED Fig. 1.
Extended Data Fig. 2 Collective invasion of breast cancer cells in 3D collagen matrix enriched with fibroblasts or fibroblast-free matrix of different density.
a, Time-dependent collective invasion of MCF-7 spheroid into LD collagen matrix. Micrographs represent 3 independent experiments. b, Collective strand length. Data represent average length of strands from n = 9 (day 1), 13 (day 3) and 13 (day 5) spheroids (3 independent experiments). c, MCF-7 cell invasion into HD collagen matrix in dependence of cocultured MRC-5 fibroblasts. Micrographs represent 3 independent experiments. d, Length of invasion strands with and without cocultured MRC-5 fibroblasts. Data without MRC-5 represent n = 15 spheroids (day 2, 4, 6) and with MRC-5, n = 13 (day 2), 31 (day 4), 39 (day 6) spheroids/condition (3 independent experiments). e, Area of collective MCF-7 sheet moving along the collagen interface of different density. Data represent n = 47 (LD), 38 (ID), 11 (HD) spheroids/condition (3 independent experiments). f, Rat-tail (RT) collagen network at 2 (LD), 4 (ID) and 6 mg/mL (HD) density, used for 3D invasion of 4T1 cells. RT, rat tail; PD, pore diameter. g, Pore diameters at different collagen densities. Data represent n = 56 (LD), 41 (ID), 26 (HD) pores/condition (3 independent experiments). h, E-cadherin localization in 4T1 cells during collective invasion in collagen with different density (48 h). Micrographs represent 3 independent experiments. i, Cell boundary/cytoplasm ratio of E-cadherin and β-catenin intensity (conditions as in h). n.d., no detached cells detectable. Data for E-cadherin represent n = 16, 19 and 18 (strands) and 21 and 9 (detached) cells for LD, ID and HD conditions, respectively, and for β-catenin 16, 19 and 18 (strands) and 16 and 6 (detached) cells for LD, ID and HD conditions, respectively (3 independent experiments). Medians (red line); dots, means from individual spheroids (b, d, e), collagen pores (g) or individual cells (i). P values, two-tailed Mann-Whitney test. Collagen matrices described in Supplementary Table 1. Scale bars, 200 µm (a, c); 50 µm (f, h). For statistical source data, see Source data for ED Fig. 2.
Extended Data Fig. 3 Collective invasion of MCF-7 cells in dense collagen matrix depends on MMP activity.
a, E-cadherin localization in collectively invading MCF-7 cells in coculture with MRC-5 fibroblasts. Micrograph representing 3 independent experiments. b, Fibroblast-generated tracks of least resistance in HD collagen containing MCF-7 strand revealed by Col 3/4 staining. Micrograph from 3 independent experiments. c, MRC-5 and HMF fibroblasts-supported invasion of MCF-7 cells into HD collagen. Micrograph from 3 independent experiments. d, MCF-7 strand length in coculture with MRC-5 and HMF fibroblasts. Median (red line); dots represent n = 34 (MRC-5) and 27 (HMF) spheroids/condition (3 independent experiments). e, MT1-MMP/MMP14 expression by MRC-5, HMF and MCF-7 cells. Western blot from 2 independent experiments. Unmodified scan of the Western blot is provided in Source data. f, Collective invasion of MCF-7 cells into HD collagen matrix after overexpression of MT1-MMP/MMP14. Insert, parental MCF-7 cell spheroid. g, Number of collective MCF-7 strands in HD collagen and MCF-7 cells after MT1-MMP overexpression. Data represent n = 17 (parental) and 30 (MMP14-OE) spheroids/condition (2 independent experiments). h-j, MCF-7 collective invasion into HD collagen with fibroblasts (h), LD collagen (i), and LD or ID interface assay (j) after interference with MMPs (GM6001). Micrographs in (h-j) from 3 independent experiments. Length of MCF-7 strands (h, i) or sheet area (j). Data in (h-i) represent n = 71/71 (h), 44/39 (i) spheroids for DMSO/GM6001, respectively, or in (j) n = 21/32 (LD) and 16/10 (ID) spheroids for DMSO/GM6001, respectively (3 independent experiments). Median (red line), dots representing the means/spheroid (d, g, h, i, j). P values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 µm (a, b); 200 µm (c, f, h-j). For statistical source data, see Source data for ED Fig. 3.
Extended Data Fig. 4 E-cadherin downregulation in MCF-7 and 4T1 cells – impact on collective invasion in collagen matrix.
a-f, Expression of E-cadherin in MCF-7-shCDH1 (a-c) and 4T1-shCDH1 cells (d-f). Example forward and side scatter plot (FSC/SSC; a, d) for gating of morphologically intact cells (3 independent experiments). Relative fluorescence intensity in MCF-7-shCDH1 cells (b) and 4T1 cells after E-cadherin downregulation with 5 different shRNA sequences (e). Western blots in (c, f) represent 3 independent experiments. Unmodified scan of the Western blots is provided in Source data. g, Expression of adhesion molecules and EMT markers after shRNA-mediated downregulation of E-cadherin in MCF-7 and 4T1 cells. Western blots represent 2 independent experiments. Unmodified scan of the Western blots is provided in Source data. h, Number of collective strands in 4T1 spheroids in matrix with different collagen densities. Dots represent means from n = 12/10 (LD), 14/11 (ID), 15/7 (HD) spheroids for shNT/shCDH1, respectively (3 independent experiments). i, Length of 4T1 collective strands in 4T1 spheroids in different collagen density. Dots represent n = 130/10 (LD), 161/26 (ID), 107/19 (HD) strands for shNT/shCDH1, respectively (3 independent experiments). j, Collective invasion of MCF-7 cells after E-cadherin downregulation during coculture with HMF fibroblasts in HD collagen. Micrographs represent 3 independent experiments. k, MCF-7 strand length in the absence or presence of HMF fibroblasts. Dots represent the average strand length per spheroid from n = 30/29 (shNT) or 20/44 (shCDH1) spheroids for either HMF condition, respectively (3 independent experiments). l, E-cadherin surface localization in MCF-7-shNT or MCF-7-CDH1 cells migrating in LD or HD interface assay. Micrographs represent 3 independent experiments. Medians (red line) in (h, i, k). P-values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 µm (j); 50 µm (l). For statistical source data, see Source data for ED Fig. 4.
Extended Data Fig. 5 Collagen matrix confinement determines invasion mode of breast cancer cells in the presence of adhesion-perturbing anti-E-cadherin antibody.
a, Single-cell dissemination of MCF-7 cells in collagen matrix of different density in the absence or presence of blocking anti-E-cadherin mAb SHE78-7. Micrographs represent 3 independent experiments. b, Number of individualized cells under conditions in (a). Dots for 3D represent n = 11/16 (LD), 11/10 (ID), 34/34 (HD) and, for interface, n = 39/36 (LD), 22/16 (ID), 50/39 (HD) spheroids for IgG/SHE78-7, respectively, from 3 independent experiments. c, Dependence of the number of individually invading cells out of MCF-7 spheroids on the pore diameter within 3D collagen matrices with different density, with culture conditions as in (b). d, Single-cell detachment of MCF-7 cells by SHE78-7 mAb in LD collagen. Micrograph represents 3 independent experiments. e, Single-cell detachment of MCF-7 cells along collagen-ablated tracks in HD collagen. Micrograph represents 3 independent experiments. f, Number of individualized MCF-7 cells in collagen-ablated tracks. After 48 h tracks were completely filled with cells, resulting in multicellular strands irrespective of antibody condition. Data represent n = 7/18 (24 h), 7/18 (48 h) spheroids for IgG/SHE78-7, respectively pooled from 3 independent experiments. g, Combined interference using antibody SHE78-7 in MCF-7-shCDH1 cells migrating in interface assay with collagen of different density. Micrographs represent 3 independent experiments. h, Number of individualized MCF-7 cells after E-cadherin downregulation in interface assay. Data represent the means from n=20 spheroids per condition pooled from 3 independent experiments. Red line in (b, f, h), median. P values, two-tailed Mann-Whitney test. Low (LD), intermediate (ID) and high (HD) density collagen matrices are described in Supplementary Table 1. Scale bars, 100 µm (a, g); 50 µm (d, e). For statistical source data, see Source data for ED Fig. 5.
Extended Data Fig. 6 Variability of cell-cell junction composition and kinetics of carcinoma cell invasion in dependence of E-cadherin status and ECM confinement.
a, b, c, F-actin (a, b), E-cadherin and p120 catenin (c) localization in 4T1-shNT or 4T1-shCDH1 cells invading from multicellular spheroids along the interface of LD (a) or HD (b, c) collagen after 48 h. Micrographs in (a-c) represent cells from 2 independent experiments. Scale bars, 50 µm. d-f, Automated tracking of H2B-mCherry positive cell nuclei of 4T1 cells moving along the interface of LD or HD collagen (time period: 27-54 h). Spatial correlation of velocity (d), temporal correlation of velocity (e) and mean squared displacement (f) of 4T1-shNT or 4T1-shCDH1 cells at the outward edge. Lines in (d-f) represent the average of n=10 spheroids/condition pooled from 2 independent experiments. g, Particle image velocimetry (PIV) analysis of H2B-mCherry positive cell nuclei of 4T1 cells in LD or HD interface assay (time period: 43-48 h). Vorticities and velocities obtained from 4T1 cell nuclei at the invasion edge, mid-invasion zone or in the spheroid core. Data represent n=10 spheroids/condition pooled from 2 independent experiments. Box plots in (g) represent the median (mid-line), the second quartile (colored band) and the 1.5 interquartile range (whiskers). Two-sided Kolmogorov-Smirnov tests indicate that all possible comparisons are statistically significant (p < 10-6). For statistical source data, see Source data for ED Fig. 6.
Extended Data Fig. 7 Effects of E-cadherin downregulation on protein expression, tumor growth and spontaneous metastasis of MCF-7 and 4T1 cells after implantation into the mammary fat pad.
a, Collective invasion of 4T1 cells into the fibroblast-rich stroma 4 days after spheroid implantation into the mammary fat pad. Example images from 2 independent tumors. b, Pan-cytokeratin (upper panels) or cytokeratin-8 (lower panels) expression in 4T1-shNT or 4T1-shCDH1 tumors in the mammary fat pad (4 days post-implantation) or in the lungs (38 days after tumor implantation in the mammary fat pad). Micrographs represent 3 independent tumor samples (upper panel) or lungs samples from 5 mice/condition (lower panels). c, Pan-cytokeratin (upper panels) and E-cadherin or vimentin (lower panels) expression in MCF-7-shNT or MCF-7-shCDH1 tumors in the mammary fat pad (6 days post-implantation). Micrographs represent 2 (upper panel) or 1 (lower panels) tumors/condition. d, Time-dependent growth in MCF-7-shNT or MCF-7-shCDH1 primary tumors. Data display the mean ± SEM from n=5 mice per group. P values, two-tailed Mann-Whitney test. e, Number of metastatic foci within the lungs of MCF-7-shNT or MCF-7-shCDH1 tumor-bearing mice up to 38 days post-implantation in the mammary fat pad. Data represent the number of metastatic cell clusters per tissue slice (from 50-100 slices per lung) from n=5 (4T1-shNT, 4T1-shCDH1, MCF-7-shCDH1) or n=6 (MCF-7-shNT) mice/group. Red line, median. P values, two-tailed Mann-Whitney test. Scale bars, 50 µm (a, b, upper panel, c) and 25 µm (b, lower panel). f, Hypothetical jamming phase diagram for cell invasion into 3D tissue. For statistical source data, see Source data for ED Fig. 7.
Supplementary information
Supplementary Tables
Supplementary Table 1. Degree of confinement of collagen matrix conditions of 3D invasion assays shown in Fig. 2, 3 and 4 Supplementary Table 2. SiRNA and shRNA sequences used to target E-cadherin, N-cadherin, P-cadherin, α-catenin, β-catenin and p120 catenin in 4T1 and MCF-7 cells. Supplementary Table 3. The regions adapted for 2 mg/mL and 6 mg/mL collagen conditions (for particle image analysis).
Supplementary Video 1
3D reconstruction of invasive ductal carcinoma lesion in a patient sample (30 µm z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive E-cadherin staining; collagen bundles (SHG); nuclei (DAPI). The video is representative for 7 independent samples. Related to Fig. 1.
Supplementary Video 2
3D reconstruction of invasive lobular carcinoma lesion in a patient sample (60 µm z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive staining for pan-cytokeratin; collagen bundles (SHG); nuclei (DAPI). The video is representative for 3 independent samples. Related to Fig. 1.
Supplementary Video 3
In silico analysis of collective and single-cell migration modes in dependence of adhesion and extracellular matrix confinement. Time evolution of representative examples of four parameter combinations corresponding to low/high cell-cell adhesion and low/high ECM confinement. Strong cell-cell adhesion leads to correlated movement without single cell release (lower row, corresponding to Fig. 5e region 2); decreasing cell-cell adhesion leads to uncorrelated movement with single cell release if ECM confinement is weak (top left, corresponding to Fig. 5e region 1) and without single cell release if confinement is high (top right, corresponding to Fig. 5e region 3). Vectors correspond to local flux of cells. The color code indicates the local velocity correlation between nearest neighbours, with red/green corresponding to anti-correlated/correlated movement. Blue circles, single cells.
Supplementary Video 4
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 5
Time-lapse movies of the invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 6 mg/ml concentration. Bright-field and H2B-mCherry (nuclei) imaging. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 6
Automatic tracking of H2B-mCherry positive nuclei (magenta circles) of control (shNT) or shCDH1 4T1 cells invading from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. Migration tracks (yellow lines). Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 7
PIV analysis of time lapse movies. The invasion of control (shNT) or shCDH1 4T1 cells from multicellular spheroids along the interface underneath the 3D collagen matrix of 2 mg/ml or 6 mg/ml concentration. The color denotes the vorticity and the arrows indicate the direction and amplitude of the local velocity. Related to Fig. 6. The video is representative for 10 spheroids per condition from 2 independent experiments.
Supplementary Video 8
Automatic 3D tracking of H2B-mCherry/LifeAct-eGFP shNT or shCDH1 4T1 cells in vivo 4 days after multicellular spheroid implantation in the mouse mammary fat pad. Time-lapse movie representing the maximum projection of 85 μm Z-stack taken every 10 min over 140 min. Related to Fig. 7. The video is representative for 3 independent experiments per condition.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 3
Unprocessed Western blots
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 4
Unprocessed Western blots
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 7
Statistical source data
Rights and permissions
About this article
Cite this article
Ilina, O., Gritsenko, P.G., Syga, S. et al. Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion. Nat Cell Biol 22, 1103–1115 (2020). https://doi.org/10.1038/s41556-020-0552-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0552-6
This article is cited by
-
Mechanical state transitions in the regulation of tissue form and function
Nature Reviews Molecular Cell Biology (2024)
-
Microinterfaces in biopolymer-based bicontinuous hydrogels guide rapid 3D cell migration
Nature Communications (2024)
-
Adherens junctions as molecular regulators of emergent tissue mechanics
Nature Reviews Molecular Cell Biology (2024)
-
Profiling native pulmonary basement membrane stiffness using atomic force microscopy
Nature Protocols (2024)
-
Extracellular vesicle-cell adhesion molecules in tumours: biofunctions and clinical applications
Cell Communication and Signaling (2023)