Abstract
Data-driven materials science has realized a new paradigm by integrating materials domain knowledge and machine-learning (ML) techniques. However, ML-based research has often overlooked the inherent limitation in predicting unknown data: extrapolative performance, especially when dealing with small-scale experimental datasets. Here, we present a comprehensive benchmark for assessing extrapolative performance across 12 organic molecular properties. Our large-scale benchmark reveals that conventional ML models exhibit remarkable performance degradation beyond the training distribution of property range and molecular structures, particularly for small-data properties. To address this challenge, we introduce a quantum-mechanical (QM) descriptor dataset, called QMex, and an interactive linear regression (ILR), which incorporates interaction terms between QM descriptors and categorical information pertaining to molecular structures. The QMex-based ILR achieved state-of-the-art extrapolative performance while preserving its interpretability. Our benchmark results, QMex dataset, and proposed model serve as valuable assets for improving extrapolative predictions with small experimental datasets and for the discovery of novel materials/molecules that surpass existing candidates.
Similar content being viewed by others
Introduction
Materials science has greatly benefited from advancements in machine learning (ML) and deep learning (DL) techniques1,2,3,4,5,6. These techniques have revolutionized the prediction of molecular properties, leveraging traditional computational approaches, such as the group contribution (GC) method7,8,9, quantitative structure-activity/property relationship (QSAR/QSPR) method10,11,12,13, quantum mechanics (QM), and molecular dynamics (MD) calculations14,15,16,17,18,19,20,21,22,23,24,25,26. Graph neural networks (GNNs) have emerged as a promising DL-based method for property prediction by embedding molecular structures in a graph architecture27,28. Moreover, pre-trained GNNs, which employ self-supervised or transfer learning, have demonstrated the highest accuracies across various benchmarks in molecular property prediction29,30,31,32,33. ML/DL techniques continue to enhance the accuracy and speed of property prediction, serving as indispensable tools for data-driven materials science.
However, a fundamental contradiction persists in ML/DL techniques regarding their inherent extrapolation difficulty, i.e., the ability to predict beyond the available data. In molecular property prediction, two main limitations arise from the range of molecular properties and the diversity of molecular structures (see Fig. 1). The primary objective of data-driven materials exploration is to identify high-performance molecules/materials that are not yet represented in databases. Hence, ML/DL models must possess the capability to extrapolate unexplored data solely from the available data. However, materials datasets often consist of small experimental results, typically containing fewer than 500 data points34,35,36,37,38,39, which inevitably carries biases due to molecular structures and property ranges. It is crucial to determine whether ML/DL models can overcome these biases and effectively extrapolate molecular properties, even when dealing with limited data, to discover novel materials/molecules that outperform existing ones.
The interpolation task represents predictions within the available property range and molecular structures, while the extrapolation task represents predictions outside the training distribution of existing data. Plot illustrates the relationship between 1D-UMAP (Uniform Manifold Approximation and Projection90) component of molecular structures and property values in the benchmark dataset of dielectric breakdown strength (EBD). Molecules with outstanding property values potentially exist in regions distant from the existing molecular space.
Several studies have highlighted significant degradation in property prediction caused by biases in both molecular properties and structures40,41,42,43,44,45. It has been recognized that differences in material composition between training and testing data can cause a substantial decrease in prediction accuracy43,44. Furthermore, a recent finding demonstrated that even within the same database, a distribution shift of crystal structures can lead to a deterioration in prediction performance41. To address this issue, researchers have explored the applicability domains (ADs) of ML/DL models in property prediction, employing techniques such as design space quality46, UMAP-based structure mapping41, disparity in multiple ML prediction41, feature distribution clustering42,47, and active learning strategies48. While these AD designs aim to differentiate reliable interpolation data from unreliable extrapolation data, they do not significantly enhance extrapolative performance. Meanwhile, various validation methods have been proposed to improve extrapolative performance by considering structure bias44,49,50 and property bias51,52. DL techniques such as generative models37, and Neural Network Potential (NNP) models53 have also shown potential for improving extrapolation tasks. Descriptor design, encompassing quantification of molecular geometry and electrostatic potential, has been explored for its contribution in predicting catalyst-free energy beyond the training range as y-based extrapolation54. However, the generalizability of these models is limited when adapting them to other tasks in a reasonable manner. Advancing the field of materials exploration requires a comprehensive understanding of extrapolative performance using standard ML/DL approaches and molecular descriptors, along with the development of a dedicated model specifically designed for extrapolation.
This article presents a comprehensive investigation of the extrapolative performance of ML/DL models in predicting various materials properties. Our objective is to provide a valuable guide for extrapolating material-specific ground truth properties, which are values observed through experiments. This serves as a practical goal in materials design, particularly when dealing with limited experimental data available34,35,55,56. While QM-assisted ML/DL models have gained attention for small-data material properties, their extrapolation performance has not been thoroughly investigated15,16,17,19,23,24,25,26,54. Our focus is not on establishing an ML benchmark solely based on materials composition, but rather on utilizing QM and other computed descriptors to assist in achieving extrapolation with the small-data properties. A large-scale benchmark was constructed using 12 experimental datasets of organic molecular properties and different molecular descriptors for ML/DL models comparison. The benchmark outcomes reveal a significant degradation in the extrapolative performance of conventional ML/DL models, especially for small-data predictions.
Here, we propose ML prediction models that exploit a diverse set of QM descriptors, denoted as QMex. Among these models, the QMex-based Interactive Linear Regression (QMex-ILR) distinguishes itself from standard ML/DL models through the following three key aspects. Firstly, it adopts a linear regression (LR) framework to ensure interpretability and to prevent overfitting within the training range for molecular property prediction, in contrast to non-linear regression (NLR)41,49. Secondly, QMex-ILR leverages the relationship between QMex descriptors and molecular properties, leading to preserved prediction performance for untrained molecular structures by calculating additional QMex descriptors57,58,59. Lastly, QMex-ILR expands the expressive power of LR while maintaining its interpretability (extrapolative performance) through the use of interaction terms between QMex descriptors and structure-based categorical information. This addresses the limitation of conventional QM-based LR models, which rely solely on a limited set of QM descriptors. Our models enable the prediction of various molecular properties not covered by existing benchmark datasets, thereby facilitating materials design endeavors.
Results
Data and model for benchmark
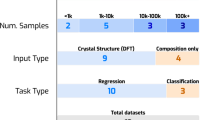
The comprehensive benchmark employed 12 datasets of fundamental and functional properties of pure organic molecules, as detailed in Table 1. All of these datasets consist of experimental data, with data points ranging from approximately 100 to 12,000, aiming at extrapolating ground truth molecular properties for small datasets. The datasets encompass a wide spectrum of physicochemical, thermal, electrical, and optical properties, which play a crucial role in the fields of chemical, environmental, agricultural, and medical sciences. Physicochemical properties have a particularly profound impact on the absorption, distribution, metabolism, excretion, and toxicity (ADMET) characteristics in drug discovery60. Numerous benchmark studies have been conducted for various ML/DL models focusing on interpolative performance15,27,31,32,61. However, their extrapolative performance remains largely unexplored. To investigate the extrapolative performance of conventional models, we utilized several datasets. These include the MoleculeNet dataset (dGs, logS, and logD)27, group contribution dataset called SPEED (logP, logS, Tb, and Tm)62, dataset by Bouteloup et al. (RI: Refractive Index)63, and Data-Warrior dataset curated by Mansouri et al. (acidic and basic pKa)64. Additionally, we incorporated two smaller-scale datasets of gas lifetime \({\log }_{10}{\tau }_{{{{\rm{life}}}}}\)38,65 and dielectric breakdown strength EBD (relative to SF6)39,66, aiming to encompass a diverse range of molecular properties. Each dataset was curated prior to training, following the procedures described in the Methods section.
We used a total of 11 ML/DL models with various molecular descriptors, including QM-based and structure-based descriptors, as outlisted in Table 2 and illustrated in Fig. 2. As ML baselines, we used Partial Least Square (PLS) regression for LR and Kernel Ridge Regression (KRR) for NLR. Molecular descriptors representing binary chemical categories, Extended-Connectivity Fingerprint (ECFP), and 2D-descriptor fingerprint (2DFP) were generated from SMILES using RDkit67, as listed in Supplementary Table 4. The chemical categories involve polarity, ionicity, and the presence of various atoms, bonds, and substructures. For DL baselines, we used two GNN models: Graph Convolutional Network (GCN)68 and Graph Isomorphism Network (GIN)69. Notably, GIN has achieved state-of-the-art (SOTA) performance on several molecular benchmarks, even without employing pre-training methods such as self-supervised or transfer learning29. To evaluate group contribution regression (GCR), we utilized the SPEED database62 and performed PLS retraining for each test. With the exception of GCR, all baseline ML/DL models employed SMILES as the raw data format, rather than 3D-coordinates.
a Summary of baseline machine learning models and molecular descriptors. Molecular structures are encoded using molecular graphs, atom groups, fingerprints, and QM descriptors, respectively. The relationships between molecular structures and properties are trained and predicted through GNNs employing Multi-Layer Perceptron (MLP) and other linear or nonlinear ML models. b Workflow of quantum mechanics-based Interactive Linear Regression (QM-ILR). Molecular structures are converted into QM descriptors and chemical categories, which are 0/1 for absence or presence of chemical features in the molecule, respectively. Interaction terms between them are generated by simple polynomial calculation considering terms for both absence and presence. PLS and Lasso regression are used to learn mixed linear correlations between QM descriptors, chemical categories, and molecular properties, as shown on the lower figures.
The QM descriptors for benchmark tests were generated using predictions from surrogate GIN models. The surrogate models were trained on an extensive dataset of density functional theory (DFT) calculations. While the ideal scenario involves performing DFT calculations for all molecules, obtaining converged results all is challenging and computationally expensive in this extensive benchmark. Consequently, we adopted surrogate GINs by obtaining DFT results for approximately half of the benchmark molecules to use as training data, as summarized in Supplementary Table 1. Note that this benchmark does not assume extrapolation capabilities of QM surrogates, and the training data for these surrogate GINs included DFT results, even for the molecules used in the benchmark test. To prevent the mixing of QM descriptors with DFT results (or surrogate training values) and surrogate test values, we unified the QM descriptors in the benchmark using the test values of the surrogate GINs. Two sets of QM descriptor datasets were used for performance comparison. One set was the QMex dataset newly calculated in this study, while the other set was extracted from the QMugs70 and CombiSolv-QM19 databases. The lists of QM descriptors and training data points for each property dataset can be found in Tables. S1–S3 in the Supplementary Information. The details for QM calculations and GNN-based QM-surrogate models are provided in the Methods section.
Evaluation procedure of interpolation and extrapolation
We used three methods for evaluating extrapolative performance, as shown in Fig. 3: (1) property range, (2) molecular structure (cluster), and (3) molecular structure (similarity). Extrapolation of property range aims to evaluate the impact of data biases resulting from a limited range of properties, commonly referred to as y-based extrapolation. Meanwhile, extrapolation of molecular structure (cluster and similarity) assesses the influence of data biases arising from limited molecular structures, employing spatial cluster mapping and molecular similarity, respectively. Each dataset was divided based on a data size for interpolation Nin of 50, 100, 200, 500, 1000, 2000, and 5000 to evaluate the impact of data size, depending on the range of molecular property, mapping of molecular structures, and range of molecular similarity. This approach enables a comprehensive evaluation of extrapolative performance across datasets of varying interpolation sizes while also considering the restricted distribution of property range or molecular structure. Note that Nin represents an estimated size due to the size fluctuations caused by the data partitioning methods. Assuming the total dataset size as Nall, the extrapolation data size (Nex) is given by Nex = Nall − Nin for the extrapolation of molecular structure, while Nex < Nall − Nin for the extrapolation of property range. The extrapolation set for property range was partitioned to ensure that the distribution of the property (y) remains outside that in the interpolation set. This involved excluding data with extrapolative inputs (x) and interpolative outputs (y) to preserve the inherent distribution of (x, y), resulting in Nex < Nall − Nin.
a Extrapolation of property range: Interpolation data consists of molecules that are closer to the mean of distribution of QM descriptors x and molecular property y. Extrapolation data consists of molecules located outside the range of y in the interpolation data. Data points other than both interpolation and extrapolation are excluded from the evaluation. b Extrapolation of molecular structure (cluster): Molecular structures are mapped with 2D-UMAP, and spectral k-means clustering is performed on UMAP distribution. Clusters that are located far away from other clusters are designated as extrapolation data. c Extrapolation of molecular structure (similarity): Molecular similarity is calculated by mean value of Tanimoto index \(\bar{{T}_{i}}\). Molecules with high similarity are included in the interpolation data, while molecules with low similarity are assigned to the extrapolation data.
After the data separation process, ensemble learning was employed for training models and testing interpolative performance using a data of size Nin. Supplementary Fig. 10 depicts a five-fold test using Nin as training and test splits to evaluate interpolative performance, while extrapolative performance was evaluated using the trained models obtained through the ensemble learning. Hence, the test set for interpolation and training set for extrapolation are identical, and Nin serves as the data size for both purposes. The hyper-parameter sets, their validated ranges, and methods for optimization for each model are listed in Supplementary Table 5. Two accuracy metrics are used: RMSE and R2. The metrics were calculated using predicted values, which were the average of predictions from 10 ensemble models. While we performed extrapolation using both molecular clustering and similarity, the following discussion focuses solely on the results of cluster extrapolation to provide a concise demonstration of structural bias. The results of similarity extrapolation exhibit similar trends, as described in Supplementary Figs. 13–24 in the Supplementary file. Detailed procedures regarding data separation, ensemble learning, and validation methods can be found in the Methods section, and the all split datasets for benchmark are provided in the available data.
Benchmark results of interpolation and extrapolation
We conducted benchmark tests on 11 models across 12 tasks, with the exception of Mol-GCR, which was tested on 4 tasks62. These tests involved 1 interpolation and 3 extrapolation tests at different splits based on Nin from 50 to 5000. Figs. 4 and 5 present representative benchmark and statistical results for interpolative and extrapolative performance. We confirmed that the predicted values finally obtained by ensemble learning sufficiently eliminate their variation, as shown in Supplementary Fig. 12. The detailed results of all tests can be found in Supplementary Figs. 13–28 in the Supplementary file. The observations from Figs. 4 and 5 are as follows: (1) In the interpolation test, NLR and GNN models have gained dominance in high-precision models (Figs. 4 and 5a). The high accuracy of QMex-NLR and 2DFP-NLR can be attributed to the utilization of critical descriptors closely associated with the target properties, such as MolLogP, MolMR, and TPSA in the 2DFP descriptors67,71, as well as several physicochemical properties in the QMex descriptors15,17,63. Conversely, our QMex-based models offer notable advantages for interpolation tasks with small-data properties, especially for Nin ≤ 500. This contrasts with the performance degradation observed in models that rely on molecular structure information, such as Mol-GIN and 2DFP-NLR. (2) The QMex-based models achieved the highest performance in both extrapolation of property range and molecular structure (cluster). They ranked among the top three on all extrapolation tasks and achieved the best performance on 19/24 tasks (Fig. 4). Notably, in the extrapolation of property, QMex-ILR and QMex-LR emerged as the best and 2nd best models on 10/12 tasks, respectively. In contrast, NLR and GNN models exhibited a substantial reduction in their regression power during the extrapolation of property range, despite achieving the best perfextrapolation of property range. The statistical analysis presented in Fig. 5b, c confirms the superiority of QMex-ILR and QMex-LR in extrapolation of property range, while QMex-ILR and QMex-NLR excel in extrapolation of molecular structure. The details of these advantages will be discussed in the following section. (3) The proposed set of QMex descriptors performs superior compared to the existing QMugs descriptors. As shown in the performance comparison in Supplementary Fig. 30, QMex-based models surpassed QMugs-based models in 79% of the benchmark tasks. The improved performance of QMex-based models can be attributed to the availability of its enhanced descriptors. For instance, QMugs lacks descriptors for charge, bond, and protonation properties, and it lacks training data for solvation energy, as indicated in Supplementary Tables 2 and 3. (4) The proposed ILR models successfully improve the performance of QM-based simple LR models. As depicted in Supplementary Fig. 31, QMex-ILR and QMugs-ILR outperformed or achieved comparable performance to QMex-LR and QMugs-LR in 96% of the interpolation tasks and 82% of the extrapolation tasks. These enhancements are also evident in Fig. 5, where QMex-ILR consistently exhibits higher scores compared to QMex-LR in the benchmarks.
The metrics were calculated using predicted values that were the average of predictions from 10 ensemble models. The numbers above bars represent rankings of model performance in the top three. The full results of extrapolation tests with RMSE and R2, including results of other Nin sizes and extrapolation of molecular structure (similarity), are provided in Supplementary Figs. 13–28 in the Supplementary Information.
The results using a interpolation scores, b extrapolation scores of property range, and c extrapolation scores of molecular structures (cluster) are shown. The interpolation scores include the results of the three extrapolation tests. The number of tasks for each size Nin changes due to variations in the data size of each property dataset.
Extrapolation for range of molecular property
Our benchmark results reveal a significant difference between the interpolation and extrapolative performance of ML/DL models. Figure 6a presents a comparison of accuracy degradation in predicting boiling point (Tb) for extrapolation of property. It is evident that the NLR and GNN models produce larger errors as the training range of the target property is confined to a smaller Nin. This observation aligns with the widely-recognized challenge of applying NLR and GNN models to distributions beyond the training range37,41,47,51. The benchmark results indicate that NLR models based on QMex or 2DFP can occasionally achieve predictions beyond the training range, owing to the presence of descriptors strongly correlated with the target property. Some instances can be observed in the benchmark results of dGs with QMex-NLR at Nin = 50 (see Supplementary Fig. 17) and RI with 2DFP-NLR at Nin = 500 (see Supplementary Fig. 22). The benchmark result also identifies that GNNs have extrapolative performance in predicting logP, logS, and Tb when trained on extensive data, as shown in the high-Tb side of Mol-GIN result in Fig. 6a. This capability is constrained to properties that tends to increase with substructure overlap, similar to group contribution, possibly due to the application of sum-pooling in the GNN architecture. We can also see that Mol-GIN and Mol-GCR lack interpolative performance at Nin = 200 with \({R}_{{{{\rm{in}}}}}^{2}\) of 0.448 and 0.223, respectively. This limitation arises from their dependency on molecular structure information for prediction, implying the inherent challenges associated with structure-based models for predicting small-data properties.
The benchmark results of extrapolation of a property range for Tb and b molecular structure (cluster) for logP are shown. The right subplots depict plots of true values versus predicted values of QMex-ILR, QMex-NLR, Mol-GIN, and Mol-GCR, which show comparison between small and large data size for interpolation (Nin). Note that the Nin serves as both the test size for interpolation and training size for extrapolation. The data points (n) and standard deviation (σtest) for interpolation and extrapolation are represented in their legends. QM-based models exhibit high performance in extrapolating molecular structures even with small datasets, in contrast to molecular structure-based models like Mol-GIN and Mol-GCR. The performance of nonlinear models like QMex-NLR and Mol-GIN, is strongly constrained by the training range of target property, while QMex-ILR overcomes this constraint, enabling extrapolation beyond the training distribution.
While NLR and GNN models have gained attention for their strong fitting capabilities and interpolative performance, they often fail to provide accurate predictions outside of the training property range. The ratio of NLR and GNN models ranking in the top three for the extrapolation of property range is only approximately 30% for Nin = 1000, 2000, and it further decreases to 10–20% when Nin ≤ 500 (Fig. 5b). The accuracy degradation can be observed in the ratio of RMSE for extrapolation to interpolation RMSEex/RMSEin, as depicted in the benchmark results of Supplementary Figs. 13–24 in the Supplementary Information. NLR and GNN models experience an approximately four- to six-fold degradation at Nin = 500 for extrapolation of property range. On the contrary, linear models can mitigate the degradation of predicted values outside the training range owing to their linearity, as evidenced by from the smaller RMSEex/RMSEin ratio compared to those of the NLR and GNN models. Mol-CGR employs linear regression but may not always be preferable for extrapolating molecular properties. It requires sufficient molecular structure information to learn group contributions to the target property, as observed from Fig. 6a. In conclusion, QM-based LR models are optimal for extrapolating properties, and in particular, QMex-ILR proves to be more suitable as it overcomes the limitations of simple LR models and existing QM descriptors.
Extrapolation for diversity of molecular structure
Figure 6b illustrates the benchmark result in the case of lipophilicity (logP) for extrapolation of molecular structure (cluster). It is observed that the extrapolative performance of structure-based models significantly depends on the number of training molecular structures, while QM-based models exhibit little dependence on the training size, when examining the extrapolation accuracy as a function of Nin in Figs. 5 and 6. The structure-based models have strong proficiency in learning the structure-property relationship with large data, whereas their efficacy declines when applied to dissimilar molecules beyond the limited training structures. The ratio of structure-based models ranking in the top three for structure extrapolation exceeds 40% for Nin ≥ 1000, while it diminishes to approximately 10–20% for Nin ≤ 500 (Fig. 5b). This trend also suggests that structure-based models have the potential to improve their extrapolative performance by training a larger set of molecular structures, while, simultaneously, that of GNNs may saturate on the redundancy in large training data72. On the contrary, the QM-based models exhibit strong extrapolative performance for molecular structures, even when trained on limited data. Although their performance tends to saturate with 200–500 points, it is sufficiently higher than or comparable level to well-trained GNNs, as shown in Fig. 6. In essence, these results emphasize the advantage of QM-based models for unfamiliar molecules that diverge from the training molecules with limited experimental data. Similarly, self-supervised GNN or transformer models hold potential in addressing this limitation by pre-training on the diversity of molecular structures29,33, although further investigations are required.
QM-based models exhibit robustness to structure bias because they do not directly learn molecular structures; rather, they capture the relationship between target properties and QM descriptors. We can preliminarily obtain QM descriptors for a variety of molecular structures using DFT calculations or well-trained surrogate GINs without the constraints of experimental data. While this benchmark employed surrogate GINs due to DFT calculations cost, we recommend the direct use of DFT results without the surrogate bias for the most accurate extrapolation in practical material discovery.
Prospects for extrapolative prediction of molecular properties
We summarize ML/DL models suitable for predicting molecular properties, as illustrated in Fig. 7. This is based on their performance in interpolation and extrapolation tasks while considering two key factors: range of molecular properties and diversity of molecular structures. In terms of interpolation, the NLR and GNN models have shown good performance owing to their high expressive capacity. It should be noted that Mol-GIN is not necessarily the optimal choice, as it requires a large training dataset to learn the diversity of molecular structure. 2DFP-NLR and QMex-NLR have high interpolative performance by utilizing pre-trained descriptors that are strongly related to the target properties. Furthermore, we have demonstrated that QMex-ILR overcomes the limitations of LR models, excelling in the interpolation of small-data properties (Fig. 5a).
For interpolation tasks, Mol-GIN and 2DFP-NLR outperform other models, while their performance can be degraded when the data size for interpolation decreases. Alternative models, such as QMex-ILR and QMex-NLR, perform better in interpolation with limited data because of the utilization of newly provided QMex descriptors. For extrapolation tasks, ML/DL models are categorized into linear/non-linear and structure-based/QM-based models. In summary, QM-based models and linear models are more suitable for overcoming the limitations of property range and molecular structure within the training data, respectively. A simple QM-based LR model faces challenges in both interpolation and extrapolative performance due to its restricted expressive power. The proposed QMex-ILR model, which incorporates interactions between the QMex descriotors and chemical category information, extends the performance of QMex-LR.
Regarding extrapolation tasks, ML/DL models can be categorized into linear/non-linear and structure-based/QM-based models to analyze their advantages and disadvantages. QM-based ILR and 2DFP-NLR fall into the category of both structure-based/QM-based models as they incorporate both structural and pre-designed physicochemical information. Our benchmark reveals the essential role of linearity in extrapolating the property range and that of QM descriptors in extrapolating molecular structures beyond training distribution, particularly in scenarios with limited training data. The structure-based models, such as Mol-GIN and Mol-GCR, have limited prediction performance as they solely rely on learning relationships between molecular structures and a target property. In contrast, QM-based models capture hidden relationships by utilizing intermediate molecular features that are pre-trained from a wide range of molecular structures. These intermediate features can be computed for any structure without any constraints from observed data, allowing QM-based models to perform well regardless of the structural diversity. As for extrapolation of property range, our benchmark results reveal the inherent limitation of QM-based NLR models. The regression performance of NLR models are highly sensitive to the training range of molecular properties, despite optimizing their regularization functions (Fig. 6a). The challenge of extrapolation can be addressed by integrating QM descriptors and linear models. Among these models, the performance of QMex-ILR stands out when compared to both existing QMugs-based and LR-based models (Figs. 5, S30, and S31). The ILR architecture enhances the expressiveness of QM-based LR model by incorporating the binary information of chemical categories to learn multiple linear relationship between QM descriptors and target properties.
This study confirms the efficacy of QMex descriptors in extrapolating a diverse set of organic molecular properties below 600 MW. The superiority of QMex, compared to Qmugs and 2DFP models, underscores the diversity of QM descriptors as a key factor contributing to better extrapolability. When we broaden the applicability of QMex-based models to encompass other properties and larger material/molecular systems, it is necessary to enhance the set of QM descriptors and chemical categories, such as transition states, molecular activity, and intermolecular interactions73,74. Particularly in the extrapolation of polymer material or biomolecular properties, it would be effective to introduce mesoscale descriptors obtained through molecular dynamics calculations and hierarchical structural information, such as chain lengths and sequence patterns, rather than focusing solely on individual molecules75,76. Our proposed method has compatibility with these strategies due to its interpretability and utilization of molecular simulation. Moreover, we investigated the possibility of improving extrapolative performance during the hyperparameter optimization using the modified cross-validation (CV) methods (Supplementary Fig. 11). The results confirm that the improvement by the CV methods is modest compared to that by the QMex-based models (see Supplementary Figs. 29 and 32). The adequacy of the QMex dataset for training surrogate GINs was also demonstrated by evaluating the impact of surrogate accuracy on benchmark performance (see Supplementary Fig. 33 and the corresponding description). These findings emphasize the robustness and reliability of our QMex-based models, particularly QMex-ILR, in delivering superior extrapolative performance to conventional ML/DL models and validation methods.
Insights and prospects into QMex-ILR
Figure 8 demonstrates an example of the superiority of QMex-ILR in learning logS(ESOL). The improvement in extrapolative performance from \({R}_{{{{\rm{ex}}}}}^{2}=0.819\) in QMex-LR to 0.896 in QMex-ILR can be attributed to the introduction of interaction terms between QM descriptors and chemical categories. Fig. 8e, f depict the relationship between QM descriptors and molecular properties, incorporating the influence of chemical category, in alignment with the feature importance. The results of feature importance with trained Lasso coefficients imply that α (polarizability) and dEo are effective in predicting logS(ESOL). In Fig. 8e, a single regression plane depicts a linear relationship between these QM descriptors and properties, revealing the limited expressiveness of QMex-LR in accurately estimating logS(ESOL) only through QM descriptors for a large number of molecules. In contrast, the improved fit with two regression planes in 8(f) highlights that QMex-ILR can learn multiple relationships between α, dEo, and logS(ESOL), considering the presence or absence of an oxygen atom. While simple LR models have their limitations in representation, ILR can capture these variances using interaction terms while maintaining interpretability. Additional results of feature importance evaluation for QMex-LR, QMex-ILR, QMugs-LR, and QMugs-ILR are shown in Supplementary Fig. 34. This analysis not only identifies QM descriptors contributing to the prediction of various molecular properties but also reveals complex linear relationships through chemical category associated with molecular structures. For instance, in the case of gas lifetime (τlife), the importance of Ehomo is substantial, and in ILR, the presence or absence of double bond is notably highlighted. This underscores the strong influence of gas reactivity with OH radicals in the troposphere, as reported in77, and highlights the variation in the influence of Ehomo on the reactivity depending on the presence of double bond.
a, b Prediction results of logS(ESOL) with PLS at Nin = 500 in extrapolation of property range. c, d Results of feature importance evaluation using trained Lasso coefficients as indicators of importance. Prior to training, multicollinearity and sparsity of features are removed, as described in the Methods section. The mean of the trained Lasso coefficients is calculated by 50 trained Lasso models through ensemble learning to eliminate coefficient fluctuations. The features with thick box-line and pink and cyan colors indicate highly important QM descriptors and interaction terms used in the following 3D-plot. The names of interaction terms represent QM descriptor, chemical category, and category presence, where 0 and 1 denote absence and presence, respectively. e, f Relationship between dEo, α (polarizability), and logS(ESOL), not considering and considering the absence or presence of an oxygen atom in molecules, where α is denoted as “alpha".
We recommend the use of PLS as a baseline model for ILR architecture due to its ability to promptly extract significant features from the interaction terms, while Lasso was specifically employed to evaluate feature importance. The interaction method can be explained by simple equations described in the Methods section. Additionally, the ILR has an architecture to optimize the inclusion or exclusion of interaction terms using a hyperparameter denoting the interaction order. Even in situations where training data is limited, QM-based ILR performs comparably to QM-based LR by excluding interaction terms to prevent overfitting during validation process.
The application of our ILR may face challenges when dealing with situations characterized by discontinuity or nonlinearity in property regression. Discontinuous relationships, commonly observed in molecular activity cliffs78, might be captured through ILR by learning multiple linear relationships using interaction terms. The ILR is designed with an architecture that allows for the incorporation of essential categorical information to address discontinuity. Even if significant categories are unclear beforehand, it can extract this critical categorical information from the results of feature interpretation. When the relationship between QM descriptors and target properties is non-linear rather than linear, feature engineering is necessary during model optimization49. This may involve introducing logarithmic functions or incorporating interactions among QM descriptors. In such cases of feature engineering, it is effective to employ specific validation methods to preserve extrapolative performance, as demonstrated in Supplementary Fig. 11.
Discussion
This paper introduced a comprehensive benchmark that evaluates the extrapolative ability of ML/DL models for molecular property prediction beyond existing datasets. The benchmark assesses the performance on 12 diverse datasets, comprising both large- and small-data properties of organic molecules. The datasets are divided into interpolation and extrapolation sets based on the range of molecular properties or structures. The results demonstrate that conventional ML/DL models experience significant deterioration in extrapolation, despite their outstanding performance in interpolation tasks. This deterioration can be attributed to the discrepancy between the distribution of property range and molecular structures in the interpolation (training) and extrapolation data. To overcome these limitations, we proposed QMex-based models that leverage a newly calculated DFT dataset called QMex, comprising 22 QM descriptors of computational results for approximately 26k molecules. The QMex-based models exhibit robustness to structure bias across various molecular properties, attributed to the enriched diversity of QMex descriptors compared to existing QM-based models. Their strength lies in leveraging the relationship between target properties and QMex descriptors without being constrained by the molecular structure of experimental data. Furthermore, our benchmark highlights the superiority of the proposed QMex-ILR to achieve extrapolation for property range beyond the training distribution. While the conventional QM-based models rely on nonlinear kernelization methods to enhance prediction performance57,58,59, these approaches struggle with extrapolation of property range. Our QMex-ILR enhances the expressive power of linear regression by incorporating interaction terms between QMex descriptors and binary information representing molecular structures.
This benchmark study highlights the critical influence of the training data size on the selection and performance of models for predicting molecular properties (Figs. 5 and 6). QMex-based models can acquire extrapolative capabilities even with a limited experimental dataset of 200–500 points, surpassing or matching the performance of well-trained GINs. The exceptional merits of QMex-based models become evident as an indispensable tool for estimating properties of unknown molecules using limited experimental data. This is also attributed to the remarkable extrapolation robustness of QMex-based models across a wide range of properties, in comparison to other accessible QM-based models. We expect our proposed model and QMex descriptors to be widely adopted in property prediction and materials design, thereby contributing substantial progress in the field of materials science.
Method
ILR architecture
A hyper-parameter d (≥−2) was introduced into the ILR to determine an existence of the interaction terms and an intersection order of categorical information. The creation of interaction terms for d≥1 and performing PLS regression can be simply implemented using PolynomialFeatures and PLSRegression functions from the scikit-learn module. This architecture highlights the strength of ILR in obtaining interpretable analytical solutions due to its simplicity. The detailed ILR architecture, including the handling of hyperparameter d, is described in the equations below.
Given that each molecule is characterized by L types QM descriptors and M types of chemical categories, as listed in Supplementary Table 4, they are represented by a vector \({{{\bf{q}}}}=({q}_{1},{q}_{2},\ldots ,{q}_{L})\in {{\mathbb{R}}}^{L}\) and a binary vector b = (b1, b2,…,bM) ∈ {0, 1}M, respectively. The existence and absence of chemical category ci in molecule molj are represented by bi as follows:
When d = −2, the ILR does not use categorical information, and its operation is the same as that of QM-based LR using L-dimensional features x:
When d = −1, binary vector b is used without interaction; instead, it is concatenated with the QM descriptors to (L + M)-dimensional features as follows:
where b expresses the existence of chemical categories.
When d ≥ 0, we consider not only the existence of chemical categories but also their absence using M-dimensional binary vector \({{{{\bf{b}}}}}^{{\prime} }\) as follows:
Through linear regression, \({{{{\bf{b}}}}}^{{\prime} }\) contributes to learning the impacts of the absence of chemical categories, while b contributes to learning the impacts of existence.
When d = 0, the binary vector (\({{{\bf{b}}}}\,{{{{\bf{b}}}}}^{{\prime} }\)) is used without interaction; instead, it is concatenated with the QM descriptors to (L + 2M)-dimensional features as follows:
When d≥1, we introduce interaction terms between QM descriptors and chemical categories. Here, the degree d influences the order of category intersection, and a M″-dimensional new binary vector b″ is represented by using bi and \({b}_{i}^{{\prime} }\) in (1) and (5) as follows:
where
When d = 1, b″ simply serves as 2M-dimensional category information representing the existence and absence of chemical categories by (b\({{{{\bf{b}}}}}^{{\prime} }\)).
After creating the new categorical information, the binary vector b″ is used to interact with QM descriptors q to (L + 1)(M″ + 1)-dimensional features x by calculating an outer product and using a row-wise vectorization as follows:
where \({b}_{j}^{{\prime\prime} }\) is an element of b″, and \({b}_{j}^{{\prime\prime} }{{{\bf{q}}}}\) serves as QM descriptors for each category.
Given whole dataset for N molecules, the matrix of QM descriptors is denoted as QN×L and that of binary categories is denoted as BN×M. The input feature matrix X is determined depending on the order d as follows:
where 1N is an N-dimensional vertical vector whose elements are all equal to 1. The element qnl corresponds to the (n, l)-th element of (Q∣1N), and bnm corresponds to the (n, m)-th element of (B″∣1N). The matrix \({{{{\bf{B}}}}}_{N\times {M}^{{\prime\prime} }}^{{\prime\prime} }\) denotes the interaction terms for N molecules, where each row corresponds to b″ in (9).
The analytical solution of QM-based ILR for estimated values of target property \(\hat{{{{\bf{y}}}}}\) can be described using feature matrix X in (11) when it is solved with PLS given by:
where (12) serves as normalization of feature matrix, Ncomp is the number of PLS components, ti refers to the PLS component vectors, and y represents the true value of the target property. Although X is composed of multidimensional interaction terms given by x in (10), PLS regression can solve the data sparsity and provide the training of the impact of each interaction term in (10) on the target property.
Data curation
Data curations were conducted on each dataset before prediction. Given that the organic molecules in the datasets were mainly distributed within a molecular weight (MW) of 600, the molecules were limited to those within 600 MW for benchmark. To remove influences of outlier data points, each property range was set within μ ± 3σ from raw data points, where μ is the mean value and σ is the standard deviation of the property range. Monoatomic molecules and molecules which cannot be converted into canonical SMILES using RDkit67 were excluded because their molecular fingerprints and graphs cannot be generated. The distribution of each property and MW after data curation is depicted in Supplementary Fig. 1.
Quantum mechanical (QM) descriptors
Supplementary Table 2 lists QMex descriptors, including data size, calculation method, and GNN-surrogate accuracy. The molecular weight (MW) and number of electrons per atom (Ne/Na) were calculated by using RDkit67. Other descriptors were calculated using the DFT in Gaussian 1679, where initial molecular structures before optimization were constructed from SMILES using openbabel. The DFT calculations include structure optimization, volume, polar, ionized energy, frequency, and self-consistent reaction field (SCRF), as well as protonated/deprotonated SCRF calculations. The protonated/deprotonated SCRF used protonated and deprotonated molecular structures determined from the following method32, where structure optimization and SCRF calculations were conducted with DFT level of B3LYP/6-31G(d). Other DFT calculations were performed with the level of B3LYP/aug-cc-pVDZ. The QMex dataset comprises 22 QM descriptors, each derived from computational results for approximately 7k–17k molecules. The molecules are selected form the benchmark datasets in Table 1 and from Pubchem database80. It covers a wide range of molecular structures within 600 MW for small-size and middle-size organic molecules compared to existing databases81,82,83,84. The total number of unduplicated molecules for training QMex descriptors is 25,650. The data distribution and correlation coefficient ρ between the calculated QMex descriptors are summarized in Supplementary Figs. 2 and 3. Notably, 96.7% of the combinations exhibit correlation coefficients within the range of ρ ≤ 0.9.
We incorporated diverse QM descriptors from existing databases, namely QMugs70 and CombiSolv-QM19, collectively referred to as QMugs descriptors in this study. The QMugs descriptors are listed in Supplementary Table 3. We extracted the calculated results of DFT (ωB97X-D/def2-SVP) from the “summary.csv" file in the QMugs database70, excluding descriptors present only in SDF files. Although the QMugs database contains calculations for 665k molecules, we randomly selected 100k molecules to reduce the computational burden of surrogate GINs. Additionally, highly correlated descriptors were pre-excluded. From the CombiSolv-QM database, we extracted the calculated results of solvation free energies in water dGw and octanol dGo obtained using the COSMOtherm software. More detailed information on the methodology can be found in19,70. The total number of unduplicated molecules for training QMugs descriptors is 106,401. The data distribution and correlation coefficient ρ between the calculated QMugs descriptors are summarized in Supplementary Figs. 4 and 5. The correlation coefficients of 91.7% are within ρ ≤ 0.9.
Surrogate model for QM descriptors
The surrogate models were implemented with GIN to learn the relationships between molecular structures and QM descriptors using a large number of DFT results. We employed multiple GINs depending on the different sets of QMex and QMugs descriptors, as detailed in Supplementary Tables 2 and 3, respectively. While the detailed evaluation procedure can be found in the subsequent sections, the surrogate GINs were trained through ensemble learning with ten iterations of a five-fold test, as shown in Supplementary Fig. 10, resulting in a total of 50 GINs for each descriptor set. We aligned the QM descriptors in the benchmark with the test values derived from these 50 trained GINs. The QM descriptors for molecules in the training set, as listed Supplementary Table 1, were predicted by 20% of the trained GINs that did not include them in the training. The QMex and QMugs descriptors for each benchmark dataset were prepared prior to the benchmark.
The evaluated results of the surrogate GINs are shown in Supplementary Figs. 6 and 7 using parity plots. The results show that surrogate GINs provide highly accurate predictions for all QMex and QMugs descriptors, with the exception of dipole moment μ. We verified that the surrogate GINs outperform or achieve comparable performance to existing state-of-the-art (SOTA) models for predicting molecular properties, such as self-supervised GNN-based MolCLRGIN29 and transformer-based MOLFORMER33, using the QM9 dataset84 (Supplementary Fig. 8).
Data separation for extrapolation test
The extrapolation of property range focuses on the evaluation of ML/DL abilities to predict molecular properties beyond the range of the training dataset. We evaluated the performance of extrapolation of the property range by dividing the distribution of the target property into the central and outer regions. However, note that dividing the data based solely on the distribution of target property y, so-called y-based split, can result in a skewed partitioning that deviates from the inherent data distribution of both features x and property y, as illustrated in Fig. 3a. To address the potential bias in data partitioning, we introduced a partitioning method that considers the distributions of both features x and the target property y. By calculating the distances from the mean values, xdis and ydis were obtained as norms \(\left\Vert {{{\boldsymbol{x}}}}-\bar{{{{\boldsymbol{x}}}}}\right\Vert\) and \(y-\bar{y}\), respectively, where \(\bar{{{{\boldsymbol{x}}}}}\) and \(\bar{y}\) represent the mean values, which are both zero as a result of standardization. Assuming the distribution of (xdis, ydis) to be \(N_{xy}((0,0),\boldsymbol{\Sigma})\) by shifting the center to (0,0), we employed a normal distribution given by \(N_{\rm{samp}}((0,0),\frac{2N_{\rm{in}}}{N_{\rm{all}}}* \boldsymbol{\Sigma })\) for weighted random sampling of 1.05Nin points from the (xdis, ydis) distribution. Here, Nin and Nall represent the sizes of the interpolation and the entire dataset, respectively. The interpolation data were extracted from the sampled data to be within ± 2σydis of the standard deviation of ydis in the sampled data, denoted as σydis. The extrapolation data included all data points outside ± 2σydis. Covariance coefficient \(\frac{2{N}_{{{{\rm{in}}}}}}{{N}_{{{{\rm{all}}}}}}\) and sampling point 1.05Nin were set to obtain interpolation data around Nin points through the random sampling with ± 2σydis extraction. The remaining data was excluded from both interpolation and extrapolation, which has extrapolative inputs (x) and interpolative output (y). Although they could contribute to testing x-basd extrapolative performance, we excluded them due to our purpose of evaluating y-based extrapolation. In the y-based extrapolation, it is advisable to exclude them because their inclusion in the training set distorts the inherent distribution of the input-output (x, y) space. To ensure alignment with the data distribution specific to each model, we divided the distribution using x solely based on the QMex descriptors.
For the extrapolation of molecular structure (cluster), we used 2D-UMAP distribution of molecular structures as illustrated in Fig. 3b. The 2D-UMAP components were calculated using n_components = 2, n_neighbors = 15, densmap = True, and dens_lambda = 5.0 as the parameter setting to extract local similarity of molecular structures while preserving density information in high-dimensional spaces of input data85. The 2D-UMAP distributions of each dataset are depicted in Supplementary Fig. 9. The input data comprised 64-dimensional PCA components of 2048-bit ECFP with radius = 2 for the molecular structures. We performed spectral k-means clustering with k = 20 to partition the 2D-UMAP distribution into 20 small clusters. We then calculated the average distance using the distance matrix between cluster centroids, with weights based on the number of samples, to evaluate the isolation score of the clusters. We extracted the clusters in ascending order of isolation, ensuring that the number of samples in each cluster fell within the range of 0.95 to 1.05 times the data size for interpolation (Nin). In cases where we were unable to secure interpolation data during a single clustering iteration, we extracted clusters larger than 1.05*Nin and subsequently performed spectral k-means clustering (with k = 20) again to eliminate any excess isolated points.
For the extrapolation of molecular structure (similarity), molecular similarity was calculated using the mean value of Tanimoto index \({\bar{T}}_{i}\), where Tanimoto index Tij represents the similarity between molecule i and molecule j86, as depicted in Figure 3c. The use of mean value \(\bar{{T}_{i}}\) allows for the assessment of global molecular similarity. Molecules with high similarity were included in the interpolation data, while molecules with low similarity were assigned to the extrapolation data based on Nin. Supplementary Fig. 9 showcases the difference between cluster and similarity extrapolation based on different structural biases. The colormap represents the molecular similarity calculated by the mean value of Tanimoto index, which aims to measure global similarity of molecular structures. Molecules with high similarity became distributed not only in dense regions but also in sparse regions, which indicates a discrepancy between the local similarity based on the density of 2D-UMAP, evaluated through the cluster method, and the global similarity evaluated by the similarity method.
Ensemble learning
Ensemble learning is important for benchmarking because it can eliminate prediction errors due to data-split biases in training/test set and training biases caused by model’s initial parameters87. The outline of ensemble learning to assess interpolative and extrapolative performance is described in Supplementary Fig. 10. The number of ensembles was set to 10 for training surrogate GINs and evaluation of interpolation and extrapolation. For each ensemble learning, we shuffled each dataset to remove its order bias and conducted five-fold test evaluation. After ensemble learning, the test accuracy of interpolation was evaluated with mean values of concatenated test sets of all ensembles. Predicted values for extrapolation and other new molecules were also calculated using all ensemble models. As a result, 50 models were used to predict a property of extrapolation data and the predicted values of 50 sets were averaged to obtain the final predicted values.
Validation method
We used four-fold cross-validation methods inside the training set to tune the hyperparameters. One of them was conventional k-fold CV, while the other three were modified with respect to the previous methods44,49,50,51,52 to enhance extrapolative performance, as shown in Supplementary Fig. 11. One of the modified methods is Bidirectional Sequential Cross-Validation (BSCV). This method aims to improve the prediction accuracy of data points that lie outside the training range, specifically for extremely low and extremely high values in the extrapolation of property range. During the five-fold test, the training data was sorted according to the target property and separated into three-fold backward CV and forward CV datasets, respectively. The validation loss was calculated using six sets of losses of backward and forward CV. For the extrapolation of molecular structure (cluster), we employed Leave One Cluster Out Cross-Validation (LOCOCV)44,49,50 using spectral k-means clustering on 2D-UMAP. This technique was expected to enhance the prediction performance outside the training distribution of molecular structures based on UMAP representation. The training data was separated into five sets of clusters for CV datasets by using spectral k-means clustering on UMAP (k = 5). The validation loss was calculated using five sets of losses. Furthermore, for the extrapolation of molecular structure (similarity), backward CV (BCV) was employed by sorting the data based on molecular similarity to improve prediction performance in datasets characterized by extremely low similarity. Data was separated into a five-fold BCV dataset, where data with high similarity was used for the training set, while data with low similarity was used for the validation set. The validation loss was calculated using five sets of losses of BCV.
Before applying these validation methods in the benchmark tasks, we evaluated their performance using BSCV, LOCOCV, and BCV for property, cluster, and similarity extrapolation, respectively, while comparing the results against k-fold CV (Supplementary Figs. 29 and 32). It is evident that LOCOCV and BCV outperformed k-fold CV in cluster extrapolation and similarity extrapolation, while k-fold CV outperformed BSCV in extrapolation of property range. This disparity in performance can be seen particularly when employing non-linear models because of the inherent limitations of non-linear regression models to predict data beyond the training range. BSCV with non-linear regression likely failed to achieve optimal hyper-parameter search, resulting in a significant decrease in performance compared to k-fold CV. Consequently, in the benchmark tasks, we adopted k-fold CV for the extrapolation of property range, LOCOCV for the extrapolation of molecular structure (cluster), and BCV for the extrapolation of molecular structure (similarity).
Feature preprocessing
After dividing the dataset into interpolation and extrapolation subsets (prior to splitting into training and test subsets), we performed feature preprocessing to address multicollinearity and sparsity in order to mitigate overfitting. Regarding multicollinearity removal, we generated a correlation matrix for the features within the interpolation data. Combinations with absolute correlation coefficients exceeding 0.9 were identified. Next, we computed the correlation between each feature in the selected combinations and the target property. Features demonstrating stronger correlation were retained, while those exhibiting weaker correlation were eliminated. The Supplementary Information includes heatmaps illustrating the correlation matrix of the QM descriptors in Supplementary Figs. 4 and 5. For sparsity removal, we calculated the most frequently occurring value within each feature using the interpolation data. Features that exhibited a predominant value accounting for more than 99% of the total values were classified as highly sparse and subsequently excluded from the feature list. Notably, in the case of group contribution regression, features solely consisting of zeros were removed owing to the presence of atom group counts over 99% of the data were zero.
Data availability
The full datasets for the benchmark and calculated results of QMex descriptors are available in Zenodo88. The other data sources are provided with this paper and Supplementary information.
Code availability
Codes for QMex-ILR, including creation of chemical categories and interaction terms and prediction test, are available at https://github.com/shimakawa-hvg/QMex-ILR. The full procedures for benchmark are provided with this paper and Supplementary information.
References
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
Elton, D. C., Boukouvalas, Z., Fuge, M. D. & Chung, P. W. Deep learning for molecular design—a review of the state of the art. Mol. Syst. Des. Eng. 4, 828–849 (2019).
Louie, S. G., Chan, Y.-H., da Jornada, F. H., Li, Z. & Qiu, D. Y. Discovering and understanding materials through computation. Nat. Mater. 20, 728–735 (2021).
Pollice, R. et al. Data-Driven strategies for accelerated materials design. Acc. Chem. Res. 54, 849–860 (2021).
Raabe, D., Mianroodi, J. R. & Neugebauer, J. Accelerating the design of compositionally complex materials via physics-informed artificial intelligence. Nat. Comput. Sci. 3, 198–209 (2023).
Westermayr, J., Gilkes, J., Barrett, R. & Maurer, R. J. High-throughput property-driven generative design of functional organic molecules. Nat. Comput. Sci. 3, 139–148 (2023).
Alshehri, A. S., Gani, R. & You, F. Deep learning and knowledge-based methods for computer-aided molecular design—toward a unified approach: State-of-the-art and future directions. Comput. Chem. Eng. 141, 107005 (2020).
Mann, V., Gani, R. & Venkatasubramanian, V. Group contribution-based property modeling for chemical product design: A perspective in the AI era. Fluid Phase Equilib. 568, 113734 (2023).
Aouichaoui, A. R. N., Fan, F., Mansouri, S. S., Abildskov, J. & Sin, G. Combining Group-Contribution concept and graph neural networks toward interpretable molecular property models. J. Chem. Inf. Model. 63, 725–744 (2023).
Miyao, T., Kaneko, H. & Funatsu, K. Inverse QSPR/QSAR analysis for chemical structure generation (from y to x). J. Chem. Inf. Model. 56, 286–299 (2016).
Baskin, I., Epshtein, A. & Ein-Eli, Y. Benchmarking machine learning methods for modeling physical properties of ionic liquids. J. Mol. Liq. 351, 118616 (2022).
Paduszyński, K., Kłȩbowski, K. & Królikowska, M. Predicting melting point of ionic liquids using QSPR approach: Literature review and new models. J. Mol. Liq. 344, 117631 (2021).
Li, R., Herreros, J. M., Tsolakis, A. & Yang, W. Machine learning-quantitative structure-property relationship (ML-QSPR) method for fuel physicochemical properties prediction of multiple fuel types. Fuel 304, 121437 (2021).
Hayashi, Y., Shiomi, J., Morikawa, J. & Yoshida, R. RadonPy: automated physical property calculation using all-atom classical molecular dynamics simulations for polymer informatics. npj Comput. Mater. 8, 1–15 (2022).
Işík, M. et al. Overview of the SAMPL6 pka challenge: evaluating small molecule microscopic and macroscopic pka predictions. J. Comput. Aided Mol. Des. 35, 131–166 (2021).
Alibakhshi, A. & Hartke, B. Improved prediction of solvation free energies by machine-learning polarizable continuum solvation model. Nat. Commun. 12, 3584 (2021).
Boobier, S., Hose, D. R. J., Blacker, A. J. & Nguyen, B. N. Machine learning with physicochemical relationships: solubility prediction in organic solvents and water. Nat. Commun. 11, 5753 (2020).
Jha, D., Gupta, V., Liao, W.-K., Choudhary, A. & Agrawal, A. Moving closer to experimental level materials property prediction using AI. Sci. Rep. 12, 11953 (2022).
Vermeire, F. H. & Green, W. H. Transfer learning for solvation free energies: From quantum chemistry to experiments. Chem. Eng. J. 418, 129307 (2021).
Ko, T. W., Finkler, J. A., Goedecker, S. & Behler, J. A fourth-generation high-dimensional neural network potential with accurate electrostatics including non-local charge transfer. Nat. Commun. 12, 398 (2021).
Noé, F., Tkatchenko, A., Müller, K.-R. & Clementi, C. Machine learning for molecular simulation. Annu. Rev. Phys. Chem. 71, 361–390 (2020).
Huang, B. & von Lilienfeld, O. A. Ab initio machine learning in chemical compound space. Chem. Rev. 121, 10001–10036 (2021).
Guan, Y. et al. Regio-selectivity prediction with a machine-learned reaction representation and on-the-fly quantum mechanical descriptors. Chem. Sci. 12, 2198–2208 (2020).
Zahrt, A. F. et al. Machine-Learning-Guided discovery of electrochemical reactions. J. Am. Chem. Soc. 144, 22599–22610 (2022).
McNaughton, A. D. et al. Machine learning models for predicting molecular UV-Vis spectra with quantum mechanical properties. J. Chem. Inf. Model. 63, 1462–1471 (2023).
Stuyver, T. & Coley, C. W. Quantum chemistry-augmented neural networks for reactivity prediction: Performance, generalizability, and explainability. J. Chem. Phys. 156, 084104 (2022).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Fung, V., Zhang, J., Juarez, E. & Sumpter, B. G. Benchmarking graph neural networks for materials chemistry. npj Comput. Mater. 7, 1–8 (2021).
Wang, Y., Wang, J., Cao, Z. & Barati Farimani, A. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4, 279–287 (2022).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Wu, J. et al. ALipSol: An Attention-Driven Mixture-of-Experts model for lipophilicity and solubility prediction. J. Chem. Inf. Model. 62, 5975–5987 (2022).
Wu, J. et al. MF-SuP-pKa: Multi-fidelity modeling with subgraph pooling mechanism for PKA prediction. Yao Xue Xue Bao (2022).
Ross, J. et al. Large-scale chemical language representations capture molecular structure and properties. Nat. Mach. Intell. 4, 1256–1264 (2022).
Zhang, Y. & Ling, C. A strategy to apply machine learning to small datasets in materials science. npj Comput. Mater. 4, 1–8 (2018).
Xu, P., Ji, X., Li, M. & Lu, W. Small data machine learning in materials science. npj Comput. Mater. 9, 1–15 (2023).
Wu, S. et al. Machine-learning-assisted discovery of polymers with high thermal conductivity using a molecular design algorithm. npj Comput. Mater. 5, 1–11 (2019).
Hatakeyama-Sato, K. & Oyaizu, K. Generative models for extrapolation prediction in materials informatics. ACS Omega 6, 14566–14574 (2021).
Hodnebrog, Ø. et al. Updated global warming potentials and radiative efficiencies of halocarbons and other weak atmospheric absorbers. Rev. Geophys. 58, e2019RG000691 (2020).
Rabie, M., Dahl, D. A., Donald, S. M. A., Reiher, M. & Franck, C. M. Predictors for gases of high electrical strength. IEEE Trans. Dielectr. Electr. Insul. 20, 856–863 (2013).
Geilhufe, R. M., Olsthoorn, B. & Balatsky, A. V. Shifting computational boundaries for complex organic materials. Nat. Phys. 17, 152–154 (2021).
Li, K., DeCost, B., Choudhary, K., Greenwood, M. & Hattrick-Simpers, J. A critical examination of robustness and generalizability of machine learning prediction of materials properties. npj Comput. Mater. 9, 1–9 (2023).
Kumagai, M. et al. Effects of data bias on machine-learning–based material discovery using experimental property data. Sci. Technol. Adv. Mater.: Methods 2, 302–309 (2022).
Stanev, V. et al. Machine learning modeling of superconducting critical temperature. npj Comput. Mater. 4, 1–14 (2018).
Meredig, B. et al. Can machine learning identify the next high-temperature superconductor? examining extrapolation performance for materials discovery. Mol. Syst. Des. Eng. 3, 819–825 (2018).
Kauwe, S. K., Graser, J., Murdock, R. & Sparks, T. D. Can machine learning find extraordinary materials? Comput. Mater. Sci. 174, 109498 (2020).
Kim, Y., Kim, E., Antono, E., Meredig, B. & Ling, J. Machine-learned metrics for predicting the likelihood of success in materials discovery. npj Comput. Mater. 6, 1–9 (2020).
Sutton, C. et al. Identifying domains of applicability of machine learning models for materials science. Nat. Commun. 11, 4428 (2020).
Lookman, T., Balachandran, P. V., Xue, D. & Yuan, R. Active learning in materials science with emphasis on adaptive sampling using uncertainties for targeted design. npj Comput. Mater. 5, 1–17 (2019).
Muckley, E. S., Saal, J. E., Meredig, B., Roper, C. S. & Martin, J. H. Interpretable models for extrapolation in scientific machine learning. Digit. Discov. 2, 1425–1435 (2023).
Durdy, S., Gaultois, M. W., Gusev, V. V., Bollegala, D. & Rosseinsky, M. J. Random projections and kernelised leave one cluster out cross validation: universal baselines and evaluation tools for supervised machine learning of material properties. Digit. Discov. 1, 763–778 (2022).
Yong, W. et al. Improving prediction accuracy of high-performance materials via modified machine learning strategy. Comput. Mater. Sci. 204, 111181 (2022).
Xiong, Z. et al. Evaluating explorative prediction power of machine learning algorithms for materials discovery using k-fold forward cross-validation. Comput. Mater. Sci. 171, 109203 (2020).
Tsubaki, M. & Mizoguchi, T. Quantum deep field: Data-Driven wave function, electron density generation, and atomization energy prediction and extrapolation with machine learning. Phys. Rev. Lett. 125, 206401 (2020).
Zahrt, A. F. et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science 363 (2019).
Gupta, V. et al. Cross-property deep transfer learning framework for enhanced predictive analytics on small materials data. Nat. Commun. 12, 6595 (2021).
Yamada, H. et al. Predicting materials properties with little data using shotgun transfer learning. ACS Cent. Sci. 5, 1717–1730 (2019).
Karelson, M., Lobanov, V. S. & Katritzky, A. R. Quantum-Chemical descriptors in QSAR/QSPR studies. Chem. Rev. 96, 1027–1044 (1996).
Puzyn, T., Suzuki, N., Haranczyk, M. & Rak, J. Calculation of quantum-mechanical descriptors for QSPR at the DFT level: is it necessary? J. Chem. Inf. Model. 48, 1174–1180 (2008).
von Lilienfeld, O. A., Müller, K.-R. & Tkatchenko, A. Exploring chemical compound space with quantum-based machine learning. Nat. Rev. Chem. 4, 347–358 (2020).
Kerns, E. H. & Di, L. (eds.) Drug-like Properties: Concepts, Structure Design and Methods (Academic Press, 2008).
Xiong, Z. et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J. Med. Chem. 63, 8749–8760 (2020).
Alshehri, A. S., Tula, A. K., You, F. & Gani, R. Next generation pure component property estimation models: With and without machine learning techniques. AIChE J. 68 (2022).
Bouteloup, R. & Mathieu, D. Improved model for the refractive index: application to potential components of ambient aerosol. Phys. Chem. Chem. Phys. 20, 22017–22026 (2018).
Mansouri, K., Grulke, C. M., Judson, R. S. & Williams, A. J. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminform. 10, 10 (2018).
Intergovernmental Panel on Climate Change (IPCC). Anthropogenic and Natural Radiative Forcing (Cambridge University Press, 2014).
Vijh, A. K. Relative electric strengths and polarizabilities of gaseous dielectrics. Mater. Chem. Phys. 12, 287–296 (1985).
Landrum, G. Rdkit: Open-source cheminformatics software (2016). https://github.com/rdkit/rdkit/releases/tag/Release_2016_09_4.
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. Preprint at https://arxiv.org/abs/1609.02907 (2016).
Xu, K., Hu, W., Leskovec, J. & Jegelka, S. How powerful are graph neural networks? Preprint at https://arxiv.org/abs/1810.00826 (2018).
Isert, C., Atz, K., Jiménez-Luna, J. & Schneider, G. QMugs, quantum mechanical properties of drug-like molecules. Sci. Data 9, 273 (2022).
Wildman, S. A. & Crippen, G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 39, 868–873 (1999).
Li, K. et al. Exploiting redundancy in large materials datasets for efficient machine learning with less data. Nat. Commun. 14, 7283 (2023).
Grambow, C. A., Pattanaik, L. & Green, W. H. Reactants, products, and transition states of elementary chemical reactions based on quantum chemistry. Sci. Data 7, 137 (2020).
Politzer, P. & Murray, J. S. The fundamental nature and role of the electrostatic potential in atoms and molecules. Theor. Chem. Acc. 108, 134–142 (2002).
Amaro, R. E. & Mulholland, A. J. Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat. Rev. Chem. 2 (2018).
Nguyen, D., Tao, L. & Li, Y. Integration of machine learning and Coarse-Grained molecular simulations for polymer materials: Physical understandings and molecular design. Front. Chem 9, 820417 (2021).
Mellouki, A., Wallington, T. J. & Chen, J. Atmospheric chemistry of oxygenated volatile organic compounds: impacts on air quality and climate. Chem. Rev. 115, 3984–4014 (2015).
van Tilborg, D., Alenicheva, A. & Grisoni, F. Exposing the limitations of molecular machine learning with activity cliffs. J. Chem. Inf. Model. 62, 5938–5951 (2022).
Frisch, M. J. et al. Gaussian 16 Revision C.01 (2016). Gaussian Inc. Wallingford CT.
Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–D1380 (2023).
Blum, L. C. & Reymond, J.-L. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J. Am. Chem. Soc. 131, 8732–8733 (2009).
Irwin, J. J. & Shoichet, B. K. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Hachmann, J. et al. The Harvard clean energy project: Large-Scale computational screening and design of organic photovoltaics on the world community grid. J. Phys. Chem. Lett. 2, 2241–2251 (2011).
Ramakrishnan, R., Dral, P. O., Rupp, M. & von Lilienfeld, O. A. Quantum chemistry structures and properties of 134-kilo molecules. Sci. Data 1, 1–7 (2014).
Narayan, A., Berger, B. & Cho, H. Assessing single-cell transcriptomic variability through density-preserving data visualization. Nat. Biotechnol. 39, 765–774 (2021).
Bajusz, D., Rácz, A. & Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 7, 20 (2015).
Zhou, Z.-H. Ensemble learning. In Zhou, Z.-H. (ed.) Machine Learning, 181–210 (Springer Singapore, Singapore, 2021).
shimakawa hvg. shimakawa-hvg/qmex-ilr: v2.1 (2023). https://doi.org/10.5281/zenodo.8180722.
Fey, M. & Lenssen, J. E. Fast graph representation learning with PyTorch geometric. Preprint at https://arxiv.org/abs/1903.02428 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861 (2018).
Acknowledgements
We thank Prof. Y. Tsuruoka and Prof. Y. Matsui (Department of Information and Communication Engineering, The University of Tokyo) for their advice and help. This work was partly achieved through the use of SQUID at the Cybermedia Center, at Osaka University.
Author information
Authors and Affiliations
Contributions
H.S. and M.S. conceived the idea. H.S. carried out the quantum chemical calculations, designed, trained and evaluated the machine learning models, and wrote the manuscript. A.K. and M.S. supervised and guided the project. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shimakawa, H., Kumada, A. & Sato, M. Extrapolative prediction of small-data molecular property using quantum mechanics-assisted machine learning. npj Comput Mater 10, 11 (2024). https://doi.org/10.1038/s41524-023-01194-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-023-01194-2