Abstract
Graph neural networks are attractive for learning properties of atomic structures thanks to their intuitive graph encoding of atoms and bonds. However, conventional encoding does not include angular information, which is critical for describing atomic arrangements in disordered systems. In this work, we extend the recently proposed ALIGNN (Atomistic Line Graph Neural Network) encoding, which incorporates bond angles, to also include dihedral angles (ALIGNN-d). This simple extension leads to a memory-efficient graph representation that captures the complete geometry of atomic structures. ALIGNN-d is applied to predict the infrared optical response of dynamically disordered Cu(II) aqua complexes, leveraging the intrinsic interpretability to elucidate the relative contributions of individual structural components. Bond and dihedral angles are found to be critical contributors to the fine structure of the absorption response, with distortions that represent transitions between more common geometries exhibiting the strongest absorption intensity. Future directions for further development of ALIGNN-d are discussed.
Similar content being viewed by others
Introduction
In materials science, graph neural networks (GNNs) have gained popularity as a surrogate model for learning properties of materials and molecular systems1,2,3,4,5,6. This popularity is partly due to the intuitive, physically informed graph encoding that represents atoms with nodes and associated bonds with edges. However, beyond atom and bond features, encoding more sophisticated structural information can be helpful or even required for accurate prediction of certain properties. For example, the bond angle information is necessary for correctly capturing electronic structure and bond hybridization7,8,9,10. Likewise, for the development of machine learning potentials where accurate energy prediction is needed, three-body (bond angle) and higher-order terms are often required to be included in the descriptor11,12,13.
One practical scenario in which three- (bond angle) and four-body (dihedral angle) interactions can be critical is spectroscopy prediction. These interactions alter the local electronic structure in ways that are often detectable in X-ray and optical absorption experiments, as well as in local chemical probes, such as nuclear magnetic resonance. The power of these experimental techniques draws in part from their sensitivity to local geometric and electronic environments; however, spectral features are often convoluted and not always straightforward to interpret or predict. This is particularly true for disordered and distorted atomic environments, which are common to interfaces and glassy systems, where slight perturbations in geometry can greatly impact resulting properties14,15,16,17.
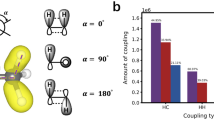
Unfortunately, conventional GNNs for atomic systems do not encode angular information. Recently, several approaches have been proposed to explicitly encode bond angles18,19,20, or bond directions from which bond angles can be implicitly retrieved21,22. Many of these were designed for non-periodic molecular structures. Alternatively, the ALIGNN approach20, which explicitly represents bond angles as edges of line graphs, provides a general formulation that is applicable to both non-periodic molecular and periodic crystal graphs4,23. However, despite its advantages, the ALIGNN encoding may not capture the full structural information of a local geometric environment. This limitation is demonstrated by the example shown in Fig. 1.
In this work, we expand the ALIGNN encoding to include the dihedral angle information. This enhanced graph representation, named ALIGNN-d, provides not only more complete structural information and but also greater interpretability. To demonstrate these advantages, we train GNN models based on ALIGNN-d alongside competing graph representations to predict infrared optical absorption spectral signatures of Cu(II) aqua complexes. These complexes are optically active and known to have high absorption sensitivity to local geometry. Utilizing configurations derived from first-principles molecular dynamics simulations, we specifically probe the role of local distortion in the GNN encoding and resulting spectroscopic signatures. Based on the results, we identify two primary advantages of the ALIGNN-d representation: (1) ALIGNN-d is a compact description that leads to the same predictive accuracy as the maximally connected graph, in which all pairwise bonds are encoded, but with greater efficiency; and (2) ALIGNN-d enables an intuitive approach to model interpretability, thanks to the explicit graph representation of bond and dihedral angles.
Results
Optical response of Cu(II) aqua complexes
The capabilities of ALIGNN-d were demonstrated by predicting infrared optical absorption spectral signatures of Cu(II) aqua complexes. These systems are broadly representative of transition metal molecular complexes that absorb optically due to d-d transitions, which are leveraged in a variety of materials applications and biological processes24. Configurations were obtained from ab-initio molecular dynamics simulations (AIMD), and optical transitions were calculated using time-dependent density functional theory (TDDFT), as described in the Methods section.
Beyond the practical implications, the high sensitivity of optical properties to the local geometry of the Cu(II) aqua complexes provides an appropriate test for ALIGNN-d. This can be seen in Fig. 2, which presents the optical transitions in the infrared regime computed from TDDFT for complexes with different instantaneous coordination numbers. In these complexes, the coordination number is found to fluctuate between four and six, with fivefold and sixfold coordination as the most and least common, respectively24,25. The results clearly indicate that the infrared optical absorption is highly sensitive to the water coordination number, with little similarity between the spectral response of the sampled configurations. Moreover, complexes with the same coordination number and visibly similar atomic configurations shown in Fig. 2a, b can generate infrared absorption profiles with noticeably different peak locations and intensities, confirming that absorption in this frequency regime is also sensitive to subtle differences in the local bonding character. The physical origin of this behavior is connected to the fundamental nature of the d-d transitions, which are nominally symmetry forbidden in ideal structures but are activated by thermal distortions. We utilize these distortions and their spectral responses as our basis to test the benefits of ALIGNN-d. Further analysis of the correlation between geometry/shape information and spectral response can be found in Supplementary Information (Supplementary Fig. 1).
The central copper ion is surrounded by either (a, b) four, (c) five, or (d) six water molecules, with fivefold coordination being the most common. Slight structural perturbations (a, b) and changes in coordination (c, d) result in noticeably different shifts of the energies and intensities. The discrete spectral peaks were computed from time-dependent density functional theory (TDDFT) simulations.
Graph representations and prediction accuracy
We first introduce our workflow for encoding the molecular features and predicting the spectroscopic signatures. Summarised in Fig. 3, this involves converting the atomic structure of the Cu(II) aqua complexes into a graph representation, followed by predicting the key spectral features from GNNs. The specific outputs of this procedure are unnormalized Gaussian functions that approximate the absorption spectra of the complexes, parameterized by the mean peak position μG, spectral width σG, and intensity AG.
The different graph representations discussed in this work are shown schematically. Note that the edge distances in these visualizations do not reflect actual real space distances. The final output is a Gaussian curve parameterized by mean μG, standard deviation σG, and amplitude AG, approximating the TDDFT-calculated discrete spectral peaks. This single-peak approximation is detailed in Supplementary Information (Supplementary Fig. 2).
As a key component of the workflow, we compare different graph representations for encoding the molecular structures. First, we consider the minimally connected graph \({{{{\rm{G}}}}}_{\min }\) that encodes only the minimal number of edges representing the nearest-neighbor atomic bonds. This graph contains the least structural information, as the bond angles and dihedral angles are not implicitly included. It can also be considered as the minimal spanning tree of the Cu(II) aqua complexes.
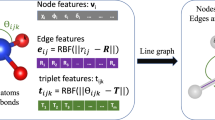
Second, at the opposite extreme, we consider the maximally connected graph \({{{{\rm{G}}}}}_{\max }\), which represents the brute-force approach that encodes all the pairwise bonds, thereby yielding complete geometric information19. Graph representations that lie between these two extremes are also considered. They are constructed using graphs defined by a specific cutoff radius, in which any pair of atoms within the cutoff distance are considered to be connected. Three values of the cutoff radius of 3, 4, and 5 Å are used, with corresponding graph representations denoted as Gc3, Gc4, and Gc5, respectively. Lastly, following the ALIGNN formulation, we incorporate angle information to \({{{{\rm{G}}}}}_{\min }\) by adding the corresponding line graph L(G), and we extend the ALIGNN formulation to explicitly represent both bond and dihedral angles in the line graph encoding, which is denoted as \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\). Since \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\) is no longer technically a line graph, it is alternatively called the dihedral graph.
ALIGNN and ALIGNN-d, alternatively expressed as \({{{{\rm{G}}}}}_{\min }\cup {{{\rm{L}}}}({{{{\rm{G}}}}}_{\min })\) and \({{{{\rm{G}}}}}_{\min }\cup {{{{\rm{L}}}}}^{\prime}({{{{\rm{G}}}}}_{\min })\), are expected to have improved representation power with respect to \({{{{\rm{G}}}}}_{\min }\). In fact, it is known that atomic numbers, bond lengths, bond angles, and dihedral angles together can fully describe the complete structure of a molecular system. This follows the principle of the Z-matrix26, which has been shown to uniquely convert this set of quantities back to the exact atomic Cartesian coordinates. Whereas ALIGNN encodes atom, bond, and bond angle features, the addition of dihedral angle information (i.e., four-body terms) in ALIGNN-d completes the Z-matrix and is therefore capable of fully describing the atomic structure. As a result, any complex geometric feature, such as distortions, chirality, and disordered configurations, can be exactly represented without explicitly including higher-order terms. In other words, ALIGNN-d implicitly has the same representation power as \({{{{\rm{G}}}}}_{\max }\), despite its considerably smaller basis.
We proceed to compare the validation performance, inference speed, and storage memory requirement based on these graph representations. It is necessary to emphasize that model memory usage during training or at inference time depends on multiple factors, such as hardware implementation, choice of graph convolution, and hyperparameters (e.g., number of channels and convolution layers). Determining such memory usage would require an individual in-depth benchmark analysis; we instead choose to compare the number of edges that fundamentally characterizes the storage memory requirement of the graphs. We also note that inclusion of line/dihedral graphs does not technically introduce additional nodes, since the nodes of line/dihedral graphs are identical to the edges of the original graph.
As expected, the inclusion of auxiliary line/dihedral graphs improves the model performance. Shown in Fig. 4, this leads to significantly improved accuracy, i.e., lower loss, over the minimum baseline \({{{{\rm{G}}}}}_{\min }\). Importantly, the validation loss of ALIGNN and ALIGNN-d lie below the curve formed by the base graphs (Fig. 4a, b), suggesting that they are more expressive relative to normal graphs when constrained to the same compute speed or memory usage. In addition, ALIGNN-d performance is noticeably better than ALIGNN, yielding a similar accuracy as \({{{{\rm{G}}}}}_{\max }\) but with significantly lower inference time (27% less) and number of edges (33% fewer), which translates to more efficient memory usage. These comparisons collectively indicate that ALIGNN-d is capable of fully describing the atomic structure with significantly faster inference speed and less memory requirement compared to \({{{{\rm{G}}}}}_{\max }\). The in-training validation losses of \({{{{\rm{G}}}}}_{\max }\), ALIGNN, and ALIGNN-d are also shown in Fig. 4c, further confirming that ALIGNN-d yields similar performance as \({{{{\rm{G}}}}}_{\max }\), and is significantly better than ALIGNN.
The validation loss, which is averaged over 8 repeated training sessions each with random initialization, is plotted with respect to a inference time, b average number of graph edges, and (c) in-training epochs. In a, the inference time is averaged over mini-batches of 64 random samples. In c, the ± 1 standard deviation of the validation loss from the repeated training sessions is shown as the semitransparent regions.
Aside from the minimal ALIGNN graph built on \({{{{\rm{G}}}}}_{\min }\), it is also informative to investigate ALIGNN representations on top of Gc3, Gc4, and Gc5 in comparison with ALIGNN-d. As expected, performance of ALIGNN generally improves when it is built on denser base graphs (Supplementary Figs. 3 and 4). However, the computational cost drastically increases to the extent that the inference compute speed is orders of magnitude higher than the original base graphs, rendering the “dense” versions of ALIGNN impractical. The same conclusion of course applies to ALIGNN-d. Nevertheless, we posit that there is no strong incentive to apply ALIGNN/ALIGNN-d to dense base graphs, as the “minimal” version of ALIGNN yields a significant performance improvement over \({{{{\rm{G}}}}}_{\min }\), and ALIGNN-d built on a minimal graph approaches the accuracy of \({{{{\rm{G}}}}}_{\max }\) while being much faster and more memory-efficient.
Finally, we validate the accuracy of the ALIGNN-d approach in predicting the infrared optical absorption spectra of the Cu(II) aqua complexes. Results presented in Fig. 5 show that our model provides accurate prediction of the mean μG and amplitude AG of the optical spectra, while yielding reasonable results for the standard deviation σG. In addition, we include in Supplementary Information a set of randomly sampled predicted peaks versus their target peaks (Supplementary Fig. 3), from which it is clear that ALIGNN-d is capable of accurately predicting a wide variety of optical absorption spectral signatures.
Discussion
Beyond efficiency and performance, ALIGNN-d can be utilized for model interpretability. Specifically, the final model output can be expressed as the sum of contributions from the individual graph components. In this way, relative contributions from the atoms, bonds, bond angles, and dihedral angles can be independently assessed. This is done by transforming the final embedding vectors (after the interaction layers shown in Fig. 3) of the atoms, bonds, and angles into non-negative scalars, which are then summed to a positive scalar final output.
We trained this interpretable variant of ALIGNN-d to predict the peak intensity AG of the infrared optical absorption spectral response of Cu(II) aqua complexes. The component-wise decomposition allows the contributions of the atomic, bond, and angular features from the model output to be directly visualized in graph format. This is illustrated in Fig. 6 for a specific aqua complex configuration, where we find that the peak intensity of optical response is primarily attributed to the central copper atom and O-Cu-O bond angles, followed by certain dihedral angles and Cu-O bonds. Other components have negligible contributions to peak intensity (note the logarithmic scale of the colorbar). As the instantaneous configurations of the aqua complex deviate from the ideal symmetry due to thermal perturbations, the individual contributions, even within the same component type, are not always equivalent. For example, some Cu-O bonds have a higher contribution than others when analyzed for a single configuration prior to thermal averaging. This also applies to the bond angles and the dihedral angles, as shown for the instantaneous complex selected in Fig. 6.
The largest contributions are derived from O-Cu-O bond angles and the central copper, followed by Cu-O bonds and certain dihedral angles. Color and line thickness indicate the magnitude of the relative contributions. For the visualizations of the bond and angle components, the atoms are faintly outlined.
This same component decomposition procedure was then applied to all configurations of the aqua complexes. The results, presented in Fig. 7, provide intuitive understanding of the physicochemical origins of the optical absorption response. As expected, the copper atoms feature high contribution relative to the oxygen and hydrogen atoms, consistent with the fact that changes in the copper’s d-shell electronic properties are largely responsible for the optical response. Nearby components that involve Cu atoms, including Cu-O bonds and O-Cu-O angles, also yield relatively higher contributions compared to others, such as water H-O-H angles. Overall, the O-Cu-O bond angles contribute significantly to the model output, consistent with physical intuition that the angular information is critical for informing electronic properties in transition metal complexes. This points to the need for explicitly incorporating angular features in the graph representation.
It is shown that contributions from dihedral angles are generally less significant than the O-Cu-O bond angles, which is expected as they represent higher-order interactions. However, they remain relatively significant when compared to features such as the hydrogen atoms, the H-O bonds, and the H-O-H angles. We therefore conclude that contributions from dihedral angles are important for resolving subtle structural differences, including those that result from weak geometric perturbations. In the spectral response, the dihedral contributions can be critical for interpreting and reproducing the fine structure.
Additional information regarding the specific coupling between geometrical distortion and the infrared optical response can be obtained from further examination of the individual distributions in Fig. 7. Specifically, the Cu-O distance, the Cu-O-H angle, and the O-Cu-O angle distributions display some degree of multimodality, suggesting that there are classes of geometries that contribute much more significantly to the optical response. To illustrate this further, we show in Fig. 8a the relationship between peak intensity contribution and bond angle for the specific case of the O-Cu-O bond angle distribution. The Cu(II) aqua complex is known to prefer octahedrally derived geometries, which is also common behavior across a range of other transition metal coordination complexes. Ideally, such geometries should exhibit angles of 90∘ and 180∘. From Fig. 8a, we determine that configurations featuring these angles are minimal contributors to peak intensity. However, as the angles are even slightly perturbed (>90∘ or <180∘), the peak intensity rapidly climbs several orders of magnitude. This is consistent with the physical understanding of the nominally symmetry-forbidden d-to-d transitions that comprise the infrared optical response of the Cu(II) aqua complex, which require thermal distortion to remove the transition constraints.
This is supported by a the relationship between O-Cu-O bond angles explored by the aqua complex during AIMD and their contributions to the peak intensity; and b the distribution of closest-matching reference geometries based on the minimum CSM value, with AIMD snapshots illustrating the most commonly explored complexes. For this analysis, only fourfold- and fivefold-coordinated complexes are considered.
Interestingly, it can be seen that high contributions to peak intensity occur for bond angles in the 120∘–140∘ range. These angles are not merely minor distortions, but rather represent new symmetries that are not octahedrally derived and have a much lower overall probability. To obtain further insight, we examined the symmetries explored during the AIMD simulations using the continuous shape measure (CSM) metric. Briefly, CSM provides a mathematically rigorous way to quantify similarity to reference geometric structures, from which closest matches to ideal symmetries can be assessed. Fig. 8b shows the breakdown of instantaneous closest-matching CSM-derived geometries exhibited during our simulation trajectories, focusing on the fivefold- and fourfold-coordinated complexes that together represent the majority of all configurations. The CSM analysis confirms that the aqua complexes prefer to adopt the square- and pyramid-like configurations, which are octahedrally derived and dominated by 90∘ and 180∘ bond angles. The fourfold-coordinated complexes also feature the seesaw configuration, which is likewise octahedrally derived. However, a significant fraction of fivefold-coordinated complexes have the closest match to a trigonal bipyramid, which is geometrically distinct and features bond angles of 120∘. These configurations are broadly reflective of the new symmetries in the 120∘–140∘ region that contribute strongly, with high certainty, to the optical response according to Fig. 8a. We therefore propose that aqua complexes that temporarily adopt new symmetries while actively transitioning from their most common geometries are critical contributors to the optical absorption in the infrared regime. In aqua complexes, such distortions occur occasionally due to thermal/solvent-induced fluctuations. However, one may imagine engineering local environments in frozen or glassy systems to bias these preferences and artificially enhance the frequency of optically responsive configurations.
In summary, our proposed graph representation ALIGNN-d represents a principled approach that efficiently captures the full geometric information of atomic structures. While the original ALIGNN paper20 focuses on predictions of general properties for crystalline materials and small-scale molecules, the center of our work is instead on disordered and distorted systems. We also show the interpretability of the ALIGNN-d approach, which was applied to elucidate contributions of specific structural features of the Cu(II) aqua complexes to their infrared optical absorption spectral signatures.
It is worth mentioning that even though ALIGNN-d is memory-efficient for capturing full structural information, requiring only sparse base graphs, ALIGNN and ALIGNN-d yield less favorable scaling as the base graph becomes dense (Supplementary Fig. 3 and 4). In this regard, ALIGNN-d performance might be less superior for highly complex materials, where a large unit cell is needed to represent the systems and therefore a dense base graph is required. Future study is therefore needed to thoroughly determine the computational cost and scalability of ALIGNN and ALIGNN-d for other classes of material and molecular systems. We also point out that in contrast to paiNN21, ALIGNN-d does not incorporate directional information and therefore cannot be used to predict tensorial properties or direction-specific properties. However, ALIGNN-d and paiNN are not necessarily mutually exclusive formulations. An interesting direction for future study is to combine angular encoding (with line graphs) and equivariant directional encoding.
Finally, we point out that our framework is general and applicable to other materials systems and properties. For instance, it could accelerate material design and selection of metal complexation with controlled shift in optical absorption for targeted optical and filtering properties. It could facilitate analysis of other spectroscopic responses in complex, disordered materials, which feature signatures that are often convoluted and difficult to interpret. It could also reveal features in the fine structure of spectra that might otherwise be overlooked. However, we caution that not all physical properties can be manifested as a simple summation of the graph components formulated in this work. In this regard, more elaborate interpretation methods may elucidate contributions of other specific structural or chemical features, opening the door to a broader range of applications.
Methods
Data preparation
Solvated Cu2+ ion was modeled using an ion in a cubic supercell with 48 water molecules at the experimental density of liquid water at ambient conditions. We carried out Car-Parrinello molecular dynamics simulations using the Quantum ESPRESSO package27, with interatomic forces derived from density functional theory (DFT) and the Perdew-Burke-Ernzerhof (PBE) exchange-correlation functional28. The interaction between valence electrons and ionic cores was represented by ultrasoft pseudopotentials29; we used a plane-wave basis set with energy cutoffs of 30 Ry and 300 Ry for the electronic wavefunction and charge density, respectively. All dynamics were run in the NVT ensemble at an elevated temperature of 380 K to correct for the overstructuring of liquid water at ambient temperatures with the PBE functional30. A time step of 8 a.u. were employed with an effective mass of 500 a.u. with hydrogen substituted with deuterium. The water was first equilibrated for 10 ps before the Cu2+ ion was inserted into the system, after which the system was equilibrated for another 10 ps. This was followed by a 40 ps production run to extract the time-averaged properties of the system.
Time dependent density functional theory (TDDFT)31 within the Tamm-Dancoff approximation32 was used to obtain the optical excitation energies of the ion complexes. Specifically, 6,846 aqua complexes were extracted roughly uniformly from the AIMD trajectory. Each complex consists of the copper ion and the surrounding water molecules within a radial cutoff of 2.92 Å, which corresponds to the first minimum of the Cu-O partial radial distribution function for the Cu2+ oxidation state. All the optical calculations were carried out using the NWChem software package33. Here, an augmented cc-pVTZ basis was used for the copper ion while an augmented cc-pVDZ basis was used for the water molecules. Four examples of the aqua complexes and their corresponding TDDFT-calculated peaks in roughly the visible-infrared range are shown in Fig. 2. The aqua complexes data was randomly partitioned into a training set of 6,161 samples and a validation set of 685 samples.
To assist with the large number of copper complexes being studied, an AiiDA34 workflow was employed to standardize and provide consistency for all calculations. AiiDA records full provenance between all calculations and ensures a robust framework for generating, storing, and analyzing results.
For GNN spectroscopy prediction, we focus on the TDDFT-calculated spectral peaks roughly in the visible-infrared range. Since the peak positions per complex tend to be close together, we approximated each complex’s discrete peaks into a single unnormalized Gaussian curve. This approximation helps simplify the output format for training a structure-to-spectrum GNN model. For example, the original spectral peaks from TDDFT may be described by a set of tuples (E1, I1), (E2, I2), . . . , where E is the peak energy, and I is the peak intensity. After the approximation, we can simply describe the spectral signature with an unnormalized Gaussian, parametrized by the mean μG, the standard deviation σG, and the amplitude AG. The single-peak approximation is further explained in Supplementary Information.
The continuous shape measure (CSM) provides a mathematically rigorous, normalized measure of the deviation of a molecular fragment geometry from an ideal reference polyhedron. As a similarity metric, CSM is bounded between 0 and 100. Low CSM value indicates high similarity to the reference shape symmetry, and thus to a highly symmetric arrangement of water molecules around the copper ion. The CSM was computed using the SHAPE code35 for the entire standard set of polyhedral reference geometries for four- and five- coordination numbers.
ALIGNN-d representation
With the ALIGNN and ALIGNN-d formulation, two graphs are used to encode one atomic structure: an original atomic graph G and its corresponding line graph L(G) or dihedral graph \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\). The nodes and edges in G represent atoms and bonds, respectively. The nodes and edges in L(G) or \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\), on the other hand, represent bonds and angles, respectively. Note that the edges in G and the nodes in L(G) or \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\) are identical entities and share the same embedding during GNN operation. Different from the original ALIGNN paper, we encoded the atomic, bond, and angular features with only the minimally required information, namely the atom type z, the bond distance d, the bond angle α, and the dihedral angle \({\alpha }^{\prime}\). In other words, in G, each node corresponds to a value of z, and each edge corresponds to a value of d. In L(G), each node also corresponds to a value of d, and each edge corresponds to a value of α. Finally, in \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\), each edge representing a dihedral angle corresponds to a value of \({\alpha }^{\prime}\). Information such as electronegativity, group number, bond type, and so on are not encoded.
We used Atomic Simulation Environment36 and PyTorch Geometric37 to construct the graph representations, and to calculate bond and dihedral angles.
Model architectures
Two different GNN model architectures were defined: one for the base graphs (\({{{{\rm{G}}}}}_{\min }\), Gc3, Gc4, Gc5, and \({{{{\rm{G}}}}}_{\max }\)); and one for ALIGNN and ALIGNN-d. We kept the two architectures as identical as we could in order to fairly evaluate and compare the model performances as a function of input graph representation. Both architectures consist of three parts: the initial encoding, the interaction operations, and the output layers (Fig. 3).
In the initial encoding, the atom type, the bond distance, and the angular values are converted from scalars to feature vectors for subsequent neural network operations. The atom type z is transformed by an Embedding layer (see PyTorch documentation38). The bond distance d is expanded into the Radial Bessel basis proposed by Klicpera et al.19. The angles α and \({\alpha }^{\prime}\) are expanded into the Gaussian basis implemented by Schnet3. However, the angular encoding treats bond angles α and dihedral angles \({\alpha }^{\prime}\) differently, and encodes their values at different channels of the expanded feature vector. Further details regarding the angular encoding are described in Supplementary Information.
The interaction operations are also known as graph convolution, aggregation, or message-passing. Following the ALIGNN paper20, we also adopted the edge-gated graph convolution39,40 for the interaction operations. The node features \({{{{\bf{h}}}}}_{i}^{l+1}\) of node i at the (l + 1)th layer is updated as
where SiLU is the Sigmoid Linear Unit activation function41; LayerNorm is the Layer Normalization operation42; Ws and Wd are weight matrices; the index j denotes the neighbor node of node i; \({\hat{{{{\bf{e}}}}}}_{ij}\) is the edge gate vector for the edge from node i to node j; and ⊙ denotes element-wise multiplication. The edge gate \({\hat{{{{\bf{e}}}}}}_{ij}^{l}\) at the lth layer is defined as
where σ is the sigmoid function, \({{{{\bf{e}}}}}_{ij}^{l}\) is the edge feature at the lth layer, and ϵ is a small constant for numerical stability. The edge features \({{{{\bf{e}}}}}_{ij}^{l}\) is updated by
where Wg is a weight matrix, and zij is the concatenated vector from the node features hi, hj, and the edge features eij:
We applied the same edge-gated convolution scheme (Eq. (1)–(3)) to operate on both the atomic graph G and the line/dihedral graphs L(G), \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\). In the case of G, the edge-gated convolution updates nodes that represent atoms, and edges that represent bonds, while exchanging information between the two, hence the term atom-bond interaction shown in Fig. 3. In the case of L(G) and \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\), the convolution updates nodes that represent bonds, and edges that represent angles, hence the term bond-angle interaction shown in Fig. 3. Note that by iteratively applying the convolution operation on both the original graph and the line/dihedral graph, the angular information stored in L(G) or \({{{\rm{{L}}}^{\prime}\rm{(G)}}}\) can propagate to G. Due to the nature of the edge-gated convolution, all the feature/embedding vectors for atoms, bonds, and angles during the interaction layers have the same length, or the same number of channels D.
Lastly, the final output layers pool (by summation) the node features of G and transform the pooled embedding into an output vector, which is a three-dimentional vector consisted of the parameters of an unnormalized Gaussian curve μG, σG, and AG. The two Linear layers have the output lengths of 64 and 3, respectively. For the interpretable variant of the model, the final output layers are replaced by a Linear layer transforming the input vectors into scalars, followed by the softplus activation \(\log (1+\exp (\cdot ))\) and global summation. This operation effectively transforms each embedding vector into a non-negative scalar before summing all the scalars into a positive scalar final output. Therefore, these component scalars can be interpreted as the atomic, bond, and angular contributions to the final output.
The model parameters for both architectures in Fig. 3 are the same, and listed in Table 1.
Model training
We used PyTorch Geometric37 to develop the GNN models. Note that although two model architectures were defined, a total of seven GNN models were trained due to the seven different graph representations studied in this work. Nonetheless, these models have the same parameters (Table 1). Similar to the ALIGNN paper20, we trained each model using the Adam optimizer43 and the 1cycle scheduler44. Each training was carried out in PyTorch38 and PyTorch Geometric37 on a NVIDIA V100 (Volta) GPU, and repeated eight times with randomly initialized weights for statistical robustness. The mean squared error (MSE) was used as the loss during training. The training parameters are the same for each training session (Table 2). All other parameters, if unspecified in this work, default to values per PyTorch 1.8.1 and PyTorch Geometric 1.7.2.
Data availability
All data required to reproduce this work is available at https://archive.materialscloud.org/record/2022.66. It can also be requested by contacting the corresponding authors.
Code availability
The source code for this work is available at https://github.com/LLNL/graphite. Alternatively, it can be requested by contacting the corresponding authors.
References
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. PMLR. 70, 1263–1272 (2017).
Coley, C. W., Barzilay, R., Green, W. H., Jaakkola, T. S. & Jensen, K. F. Convolutional embedding of attributed molecular graphs for physical property prediction. J. Chem. Inf. Model 57, 1757–1772 (2017).
Schütt, K. T., Sauceda, H. E., Kindermans, P.-J., Tkatchenko, A. & Müller, K.-R. Schnet–a deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Xie, T. & Grossman, J. C. Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys. Rev. Lett. 120, 145301 (2018).
Yang, K. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019).
Chen, C., Ye, W., Zuo, Y., Zheng, C. & Ong, S. P. Graph networks as a universal machine learning framework for molecules and crystals. Chem. Mater. 31, 3564–3572 (2019).
Linker, G.-J., van Duijnen, P. T. & Broer, R. Understanding trends in molecular bond angles. J. Phys. Chem. A 124, 1306–1311 (2020).
Timoshenko, J. & Frenkel, A. I. "inverting” x-ray absorption spectra of catalysts by machine learning in search for activity descriptors. ACS Catal. 9, 10192–10211 (2019).
Guda, A. et al. Machine learning approaches to xanes spectra for quantitative 3d structural determination: The case of co2 adsorption on cpo-27-ni mof. Radiat. Phys. Chem. 175, 108430 (2020).
Guda, A. A. et al. Quantitative structural determination of active sites from in situ and operando xanes spectra: from standard ab initio simulations to chemometric and machine learning approaches. Catal Today 336, 3–21 (2019).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett 98, 146401 (2007).
Samanta, A. Representing local atomic environment using descriptors based on local correlations. J. Chem. Phys. 149, 244102 (2018).
Lindsey, R. K., Fried, L. E. & Goldman, N. Chimes: A force matched potential with explicit three-body interactions for molten carbon. J. Chem. Theory Comput. 13, 6222–6229 (2017).
Pham, T. A. et al. Integrating ab initio simulations and x-ray photoelectron spectroscopy: Toward a realistic description of oxidized solid/liquid interfaces. J. Phys. Chem. Lett. 9, 194–203 (2018).
Velasco-Velez, J.-J. et al. The structure of interfacial water on gold electrodes studied by x-ray absorption spectroscopy. Science 346, 831–834 (2014).
Pham, T. A. et al. Electronic structure of aqueous solutions: Bridging the gap between theory and experiments. Sci. Adv. 3, e1603210 (2017).
Wan, L. F. & Prendergast, D. The solvation structure of mg ions in dichloro complex solutions from first-principles molecular dynamics and simulated x-ray absorption spectra. J. Am. Chem. Soc. 136, 14456–14464 (2014).
Park, C. W. & Wolverton, C. Developing an improved crystal graph convolutional neural network framework for accelerated materials discovery. Phys. Rev. Mater. 4, 063801 (2020).
Klicpera, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. Preprint at https://arxiv.org/abs/2003.03123 (2020).
Choudhary, K. & DeCost, B. Atomistic line graph neural network for improved materials property predictions. npj Comput Mater 7, 1–8 (2021).
Schütt, K., Unke, O. & Gastegger, M. Equivariant message passing for the prediction of tensorial properties and molecular spectra. PMLR 139, 9377–9388 (2021).
Chapman, J., Batra, R. & Ramprasad, R. Machine learning models for the prediction of energy, forces, and stresses for platinum. Comput. Mater. Sci. 174, 109483 (2020).
Chapman, J., Goldman, N. & Wood, B. C. Efficient and universal characterization of atomic structures through a topological graph order parameter. npj Comput. Mater. 8, 37 (2022).
Qiu, S. R. et al. Origins of optical absorption characteristics of cu 2+ complexes in aqueous solutions. Phys. Chem. Chem. Phys. 17, 18913–18923 (2015).
Pasquarello, A. et al. First solvation shell of the cu (ii) aqua ion: evidence for fivefold coordination. Science 291, 856–859 (2001).
Parsons, J., Holmes, J. B., Rojas, J. M., Tsai, J. & Strauss, C. E. Practical conversion from torsion space to cartesian space for in silico protein synthesis. J. Comput. Chem. 26, 1063–1068 (2005).
Giannozzi, P. et al. Quantum espresso: a modular and open-source software project for quantum simulations of materials. J. Phys. Condens. Matter. 21, 395502 (2009).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865 (1996).
Vanderbilt, D. Soft self-consistent pseudopotentials in a generalized eigenvalue formalism. Phys. Rev. B 41, 7892 (1990).
Grossman, J. C., Schwegler, E., Draeger, E. W., Gygi, F. & Galli, G. Towards an assessment of the accuracy of density functional theory for first principles simulations of water. J. Chem. Phys. 120, 300–311 (2004).
Runge, E. & Gross, E. K. Density-functional theory for time-dependent systems. Phys. Rev. Lett. 52, 997 (1984).
Hirata, S. & Head-Gordon, M. Time-dependent density functional theory within the tamm–dancoff approximation. Chem. Phys. Lett. 314, 291–299 (1999).
Apra, E. et al. Nwchem: Past, present, and future. J. Chem. Phys. 152, 184102 (2020).
Huber, S. P. et al. Aiida 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance. Sci. Data 7, 1–18 (2020).
Casanova, D. et al. Minimal distortion pathways in polyhedral rearrangements. J. Am. Chem. Soc. 126, 1755–1763 (2004).
Larsen, A. H. et al. The atomic simulation environment-a python library for working with atoms. J. Phys. Condens. Matter. 29, 273002 (2017).
Fey, M. & Lenssen, J. E. Fast graph representation learning with pytorch geometric. Preprint at https://arxiv.org/abs/1903.02428 (2019).
Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems 32, 8026–8037 (2019).
Bresson, X. & Laurent, T. Residual gated graph convnets. Preprint at https://arxiv.org/abs/1711.07553 (2017) .
Dwivedi, V. P., Joshi, C. K., Laurent, T., Bengio, Y. & Bresson, X. Benchmarking graph neural networks. Preprint at https://arxiv.org/abs/2003.00982 (2020).
Elfwing, S., Uchibe, E. & Doya, K. Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw 107, 3–11 (2018).
Ba, J. L., Kiros, J. R. & Hinton, G. E. Layer normalization. Preprint at https://arxiv.org/abs/1607.06450 (2016).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Smith, L. N. & Topin, N. Super-convergence: Very fast training of neural networks using large learning rates. Artificial Intelligence and Machine Learning for Multi-Domain Operations Applications 11006, 1100612 (2019).
Acknowledgements
The authors are partially supported by the Laboratory Directed Research and Development (LDRD) program (20-SI-004) at Lawrence Livermore National Laboratory. This work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract No. DE-AC52-07NA27344.
Author information
Authors and Affiliations
Contributions
T.A.P., S.R.Q., X.C., and B.C.W. supervised the research. B.C.W. computed the MD trajectory of the solvated copper ion. N.K. developed the automated Aiida workflow for TDDFT calculations, with assistance from S.W. T.H. performed the TDDFT calculations, developed the ALIGNN-d representation, and trained the GNN models. T.H., T.A.P., and B.C.W. wrote the manuscript with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hsu, T., Pham, T.A., Keilbart, N. et al. Efficient and interpretable graph network representation for angle-dependent properties applied to optical spectroscopy. npj Comput Mater 8, 151 (2022). https://doi.org/10.1038/s41524-022-00841-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-022-00841-4
This article is cited by
-
Quantifying disorder one atom at a time using an interpretable graph neural network paradigm
Nature Communications (2023)
-
Graph deep learning accelerated efficient crystal structure search and feature extraction
npj Computational Materials (2023)
-
Deep contrastive learning of molecular conformation for efficient property prediction
Nature Computational Science (2023)
-
Materials fatigue prediction using graph neural networks on microstructure representations
Scientific Reports (2023)
-
Explainable machine learning in materials science
npj Computational Materials (2022)