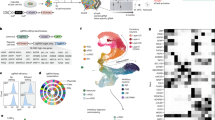
Abstract
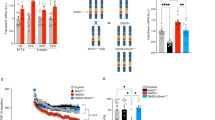
ARID1A, an SWI/SNF chromatin-remodeling gene, is commonly mutated in cancer and hypothesized to be a tumor suppressor. Recently, loss-of-function of ARID1A gene has been shown to cause intellectual disability. Here we generate Arid1a conditional knockout mice and investigate Arid1a function in the hippocampus. Disruption of Arid1a in mouse forebrain significantly decreases neural stem/progenitor cells (NSPCs) proliferation and differentiation to neurons within the dentate gyrus (DG), increasing perinatal and postnatal apoptosis, leading to reduced hippocampus size. Moreover, we perform single-cell RNA sequencing (scRNA-seq) to investigate cellular heterogeneity and reveal that Arid1a is necessary for the maintenance of the DG progenitor pool and survival of post-mitotic neurons. Transcriptome and ChIP-seq analysis data demonstrate that ARID1A specifically regulates Prox1 by altering the levels of histone modifications. Overexpression of downstream target Prox1 can rescue proliferation and differentiation defects of NSPCs caused by Arid1a deletion. Overall, our results demonstrate a critical role for Arid1a in the development of the hippocampus and may also provide insight into the genetic basis of intellectual disabilities such as Coffin–Siris syndrome, which is caused by germ-line mutations or microduplication of Arid1a.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The scRNA-seq data used in this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences with accession number GSA: CRA010586. The ChIP-seq data used in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE169562.
References
Bartsocas CS, Tsiantos AK. Mental retardation with absent fifth fingernail and terminal phalanx. Am J Dis Child. 1970;120:493–4.
Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–8.
Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92.
Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304.
Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95.
Diana CH, Gerald RC. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420.
Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–80.
Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601.
Takeda T, Banno K, Okawa R, Yanokura M, Iijima M, Irie-Kunitomi H, et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol Rep. 2016;35:607–13.
Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4.
Wu JN, Roberts CWM. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43.
Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, et al. Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell. 2019;35:177–190.e178.
Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302.
Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet. 2020;52:187–97.
Bidart M, El Atifi M, Miladi S, Rendu J, Satre V, Ray PF, et al. Microduplication of the ARID1A gene causes intellectual disability with recognizable syndromic features. Genet Med. 2017;19:701–10.
Gao XL, Tate P, Hu P, Tjian R, Skarnes WC, Wang ZES. cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–61.
Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C. 2014;166:333–49.
Goncalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: from stem. Cells Behav Cell. 2016;167:897–914.
Miller SM, Sahay A. Functions of adult-born neurons in hippocampal memory interference and indexing. Nat Neurosci. 2019;22:1565–75.
Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–44.
Liu PP, Tang GB, Xu YJ, Zeng YQ, Zhang SF, Du HZ, et al. MiR-203 interplays with polycomb repressive complexes to regulate the proliferation of neural stem/progenitor cells. Stem Cell Rep. 2017;9:190–202.
Lei X, Jiao J. UTX affects neural stem cell proliferation and differentiation through PTEN signaling. Stem Cell Rep. 2018;10:1193–207.
Liu X, Dai SK, Liu PP, Liu CM. Arid1a regulates neural stem/progenitor cell proliferation and differentiation during cortical development. Cell Prolif. 2021;54:e13124.
Han L, Madan V, Mayakonda A, Dakle P, Woon TW, Shyamsunder P, et al. Chromatin remodeling mediated by ARID1A is indispensable for normal hematopoiesis in mice. Leukemia. 2019;33:2291–305.
Abdelrahim EA, Eltony SA. Postnatal development of the hippocampal formation in male albino rats. Egypt J Histol. 2011;34:346–64.
Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–57.
Lavado A, Lagutin OV, Chow LML, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8:16.
Yu Z, Lin D, Zhong Y, Luo B, Liu S, Fei E, et al. Transmembrane protein 108 involves in adult neurogenesis in the hippocampal dentate gyrus. Cell Biosci. 2019;9:9.
Zhong SJ, Ding WY, Sun L, Lu YF, Dong H, Fan XY, et al. Decoding the development of the human hippocampus. Nature. 2020;577:531.
Hainmueller T, Bartos M. Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat Rev Neurosci. 2020;21:153–68.
Schrier SA, Bodurtha JN, Burton B, Chudley AE, Chiong MAD, D'Avanzo MG, et al. The Coffin-Siris syndrome: A proposed diagnostic approach and assessment of 15 overlapping cases. Am J Med Genet Part A. 2012;158A:1865–76.
Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–8.
Barbelanne M, Tsang WY. Molecular and cellular basis of autosomal recessive primary microcephaly. BioMed Res Int. 2014;2014:547986.
Kosho T, Miyake N, Carey JC. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am J Med Genet C Semin Med Genet. 2014;166C:241–51.
Bilha SC, Teodoriu L, Velicescu C, Caba L. Pituitary hypoplasia and growth hormone deficiency in a patient with Coffin-Siris syndrome and severe short stature: case report and literature review. Arch Clin Cases. 2022;9:121–5.
Barish S, Barakat TS, Michel BC, Mashtalir N, Phillips JB, Valencia AM, et al. BICRA, a SWI/SNF complex member, is associated with BAF-disorder related phenotypes in humans and model organisms. Am J Hum Genet. 2020;107:1096–112.
Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–95.
Matsumoto S, Banine F, Struve J, Xing RB, Adams C, Liu Y, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–83.
Stergiopoulos A, Elkouris M, Politis PK. Prospero-related homeobox 1 (Prox1) at the crossroads of diverse pathways during adult neural fate specification. Front Cell Neurosci 2014;8:454.
Shang XY, Shi Y, He DD, Wang L, Luo Q, Deng CH, et al. ARID1A deficiency weakens BRG1-RAD21 interaction that jeopardizes chromatin compactness and drives liver cancer cell metastasis. Cell Death Dis. 2021;12:990.
Wang Z, Chen K, Jia Y, Chuang JC, Sun X, Lin YH, et al. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nat Cancer. 2020;1:909–22.
Xu P, Xu J, Li Z, Yang Z. Expression of TRPC6 in renal cortex and hippocampus of mouse during postnatal development. PLoS ONE. 2012;7:e38503.
Ciric T, Cahill SP, Snyder JS. Dentate gyrus neurons that are born at the peak of development, but not before or after, die in adulthood. Brain Behav. 2019;9:e01435.
Li GN, Pleasure SJ. Morphogenesis of the dentate gyrus: What we are learning from mouse mutants. Dev Neurosci. 2005;27:93–99.
Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci USA. 2002;99:7090–5.
Bagri A, Gurney T, He XP, Zou YR, Littman DR, Tessier-Lavigne M, et al. The chemokine SDF1 regulates migration of dentate granule cells. Development. 2002;129:4249–60.
Armand EJ, Li J, Xie F, Luo C, Mukamel EA. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron. 2021;109:11–26.
Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife. 2017;6:e30506
Trizzino M, Barbieri E, Petracovici A, Wu S, Welsh SA, Owens TA, et al. The tumor suppressor ARID1A controls global transcription via pausing of RNA polymerase II. Cell Rep. 2018;23:3933–45.
Beique J-C, Lin D-T, Kang M-G, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci USA. 2006;103:19535–40.
Liu PP, Xu YJ, Dai SK, Du HZ, Wang YY, Li XG, et al. Polycomb protein EED regulates neuronal differentiation through targeting SOX11 in hippocampal dentate gyrus. Stem Cell Rep. 2019;13:115–31.
Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–9.
Liu PP, Dai SK, Mi TW, Tang GB, Wang Z, Wang H, et al. Acetate supplementation restores cognitive deficits caused by ARID1A haploinsufficiency in excitatory neurons. EMBO Mol Med. 2022;14:e15795.
Doyle DZ, Lam MM, Qalieh A, Qalieh Y, Sorel A, Funk OH, et al. Chromatin remodeler Arid1a regulates subplate neuron identity and wiring of cortical connectivity. Proc Natl Acad Sci USA. 2021;118.
Liu J, Liu S, Gao H, Han L, Chu X, Sheng Y, et al. Genome-wide studies reveal the essential and opposite roles of ARID1A in controlling human cardiogenesis and neurogenesis from pluripotent stem cells. Genome Biol. 2020;21:169.
Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol. 2017;19:962–73.
Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–8.
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62.
Zhang M, Chen D, Xia J, Han W, Cui X, Neuenkirchen N, et al. Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev 2017;31:1354–69.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Feng JX, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40.
Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Ramírez F, Ryan D, Grüning B, Bhardwaj V, Kilpert F, Richter A, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165.
Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Acknowledgements
The authors gratefully acknowledge and thank Dr. Zhong Wang at the University of Michigan for providing Arid1aflox/flox mice.
Funding
This work was supported by grants from the National Key Research and Development Program of China Project (2021YFA1101402), the Informatization Plan of Chinese Academy of Sciences (CAS-WX2021SF-0301), and the National Science Foundation of China (82271428), and the Open Project Program of State Key Laboratory of Stem Cell and Reproductive Biology.
Author information
Authors and Affiliations
Contributions
P-PL: Conceptualization; formal analysis; validation; methodology; writing-original draft; approval of draft; S-PL: Data curation; methodology; validation; approval of draft. XLi: Formal analysis; methodology; validation; G-BT: Conceptualization; formal analysis, writing-original draft; XLiu: Data curation; Formal analysis; validation. S-KD: Formal analysis; validation; L-FJ: Formal analysis; validation; X-WL: Formal analysis; validation; X-GL: approval of draft; B-YH: re-analyze seq-data; approval of draft; J-WJ: re-analyze seq-data; approval of draft; Z-QT: Data curation; formal analysis and interpretation; funding acquisition; approval of draft. C-SH: Conceptualization; Project administration. C-ML: Conception and design; data curation; formal analysis and interpretation; supervision; funding acquisition; approval of draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Our studies did not include human participants, human data or human tissue.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, PP., Lu, SP., Li, X. et al. Abnormal chromatin remodeling caused by ARID1A deletion leads to malformation of the dentate gyrus. Cell Death Differ 30, 2187–2199 (2023). https://doi.org/10.1038/s41418-023-01199-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01199-w
This article is cited by
-
Neuropathic pain development following nerve injury is mediated by SOX11-ARID1A-SOCS3 transcriptional regulation in the spinal cord
Molecular Biology Reports (2024)