Abstract
The microtubule network is crucial for cell structure and function. Patronin is a conserved protein involved in protecting the minus end of microtubules. Conversely, Klp10A is a kinesin-like microtubule depolymerase. Here we report the role of Drosophila Patronin and Klp10A for cell survival in developing organs. Loss of Patronin reduces the size of organs by activation of a caspase in imaginal discs. Reduced wing by Patronin RNAi is suppressed by knockdown of Spastin (Spas) but not Katanin 60, suggesting that Patronin is inhibitory to the severing function of Spas at the minus end. Patronin RNAi phenotype is also recovered by overexpressing Death-associated inhibitor of apoptosis 1 (Diap1), a Yorkie target gene. Heterozygote mutations in Hippo pathway genes, including hippo and warts (wts), suppress the Patronin RNAi wing phenotypes. Furthermore, Patronin physically interacts with Merlin and Expanded while reducing their function. Patronin and Klp10A antagonistically regulate their levels. Wing phenotypes of Patronin RNAi are rescued by knockdown of Klp10A, consistent with their antagonistic interaction. Klp10A overexpression also causes organ size reduction that is partially suppressed by Diap1 overexpression or wts heterozygote mutation. Taken together, this study suggests that the antagonistic interaction between Patronin and Klp10A is required for controlling cell survival and organ size by modulating microtubule stability and Hippo components.
Similar content being viewed by others
Introduction
Microtubules are crucial for cell structure and function. Microtubules have polarized structures with plus and minus ends and are dynamically regulated for its growth or depolymerization. Protection of the minus end is important for the maintenance of microtubules [1,2,3,4,5]. Drosophila Patronin, also known as Short spindle phenotype 4 (Ssp4), is a microtubule-binding protein that stabilizes the minus end [6, 7]. Patronin protects the minus end by antagonizing Klp10A, a kinesin-13 family protein involved in the depolymerization of microtubules at the minus end [7,8,9,10,11].
Recent studies in Drosophila have shown that Patronin is required for assembling non-centrosomal microtubule (ncMT) organizing center in the oocyte [12, 13]. Patronin interacts with the spectraplakin protein Short stop (Shot) and localizes apically in ovarian follicle cells to form apical–basal microtubule arrays [14]. Mammalian Patronin homologs, CAMSAP2/3, are essential for orienting the apical–basal polarity of ncMT in epithelial cells [15, 16]. However, little is known whether Patronin/CAMSAP family proteins are involved in the regulation of growth and cell survival signaling in epithelial tissues.
Regulation of organ size depends on signaling mechanisms that control growth, proliferation, and survival of cells. The Hippo pathway is a conserved mechanism for the regulation of organ size. The Hpo–Wts kinase cascade negatively regulates the downstream pathway by phosphorylating Yorkie (Yki) transcriptional coactivator [17]. Inhibition of Yki phosphorylation allows the nuclear entry of Yki [18,19,20]. Nuclear Yki can activate the expression of growth-promoting genes, including the cell cycle gene Cyclin E (CycE) and the antiapoptotic gene Death-associated inhibitor of apoptosis 1 (Diap1) [17]. The Hippo kinase cascade is regulated by a protein complex of the FERM domain proteins, Merlin (Mer) and Expanded (Ex), and the WW domain protein Kibra [21, 22]. These upstream factors are required for the activation of Hpo, thus inhibiting the Yki function.
Hippo signaling is also regulated by actin cytoskeletal factors, including capping proteins and cofilin [23,24,25]. In contrast, the role of microtubules in Hippo signaling in vivo has been rarely studied. NF2, the mammalian Mer homolog, can bind and stabilize microtubules [26,27,28]. Mer is also reported to move along microtubules and its mobility affects Yki localization [29]. However, it remains to be determined whether microtubule stability is required for growth or cell death regulation by affecting the Hippo pathway.
In this study, we provide evidence that organ size is regulated by the antagonistic interaction between Patronin and Klp10A in Drosophila. We show that apoptosis and reduced organ size by loss of Patronin can be restored by reducing Hippo signaling, implying a link between Patronin and Hippo signaling.
Materials and methods
Fly stocks and genetics
Drosophila melanogaster stocks were cultured in a standard cornmeal medium. Most genetic crosses were carried out at 25 °C unless stated otherwise. Females were used for the analysis of wing and eye size, although no significant phenotypic difference was detected between males and females. Third-instar larvae were used for immunostaining of imaginal discs.
w1118 was used as the wild-type control. GAL4 lines were as follows: en-GAL4, nub-GAL4, ey-GAL4, ptc-GAL4, and tub-GAL4 (Kyoto 108069). RNAi lines were: Patronin RNAi (Bloomington Drosophila Stock Center: BDSC 36659; Vienna Drosophila Resource Center: VDRC v27654 and v108927), Mer RNAi (VDRC v7161 and BDSC 28007), bnl RNAi (BDSC 34572 and NIG 4608R-2), Klp10A RNAi (VDRC v41534 and BDSC 3396), spas RNAi (BDSC 53331 and 27570), and Kat60 RNAi (VDRC v106487 and BDSC 28375). UAS lines were: UAS-Diap1 (BDSC 6657), UAS-CycE (Kyoto 107876), UAS-yki-V5 (BDSC 28819), UAS-p35 (BDSC 5072), UAS-Eb1-EGFP (BDSC 36861), UAS-GFP (2nd), UAS-GFPNLS (3rd), UAS-Dcr2 (1st), and UAS-Dcr2 (3rd) (BDSC 24651). Mutant lines were: PatroninEY05252 (BDSC 16647, Kyoto 114436), PatroninK07433 (BDSC 10672), hpoKS240 (BDSC 25085), wtsP2 [30], exBQ (BDSC 39689), ex697 (BDSC 44248), and exe1 (BDSC 44249). Other lines were: y w ey-flp, P{5xgIBS-lacZ.38–1}TPN1; FRT42D (BDSC 5616), hs-flp; FRT 42D Ubi-GFPNLS (gift from Dr. Georg Halder), FRT 42D (BDSC 1802), Patronin:YFP (gift from Dr. Daniel St Johnston [12]), Df(2R)BSC355 (BDSC 24379), Ubi-p63E-Patronin A-GFP (BDSC 55128), Δ2–3, balancers, and Diap1-lacZ (gift from Dr. Jongkyeong Chung). According to VDRC, Patronin RNAiKK100898 has a predicted off-target to the branchless (bnl) gene. However, two independent bnl RNAi lines do not cause any change in the wing (Fig. S2h–m, o, p).
Antibodies, immunostaining, and imaging
Third-instar larvae were dissected in cold phosphate-buffered saline (PBS) (pH 7.4). Samples were fixed in PLP (2–4% paraformaldehyde, 10 mM sodium periodate, 75 mM lysine, and 35 mM sodium phosphate buffer or PBS, pH 7.4) for 15 min at room temperature (RT). Samples were washed with PBS for 5 min two times. Samples were treated with blocking buffer (0.3% Triton X-100, 5% normal goat serum, and 0.08% NaN3 in PBS, or 0.3% Triton X-100, 0.5% BSA, 0.01% NaN3 in PBS) for 2 h at 4 °C or 40 min at RT, and incubated with primary antibodies diluted in washing buffer (0.3% Triton X-100 in PBS) overnight at 4 °C. Primary antibodies were: rabbit anti-cleaved Death caspase-1 (Dcp-1) (Asp216) (Cell Signaling Technology 9578, 1:100), sheep anti-GFP (Bio-Rad 4745–1051, 1:200), chicken anti-GFP (Abcam 13970, 1:500), rabbit anti-Patronin (gift from Dr. Emmanuel Derivery, 1:100 [31]), rabbit anti-Patronin (this study, 1:100), rabbit anti-Yki antiserum (gift from Dr. Kenneth Irvine, 1:500 [19]), rabbit anti-Klp10A (gift from Dr. David Glover, 1:200 [32]), guinea pig anti-Mer antiserum (gift from Dr. Richard Fehon, 1:2000 [33]), mouse anti-Armadillo (Arm) (DSHB N2 7A1, 1:200), rat anti-Ci (DSHB 2A1, 1:100), chicken anti-β-Gal (Abcam 9631, 1:50), rabbit anti-Phospho-Histone H3 (PH3) (Ser 10, Merck 06–570, 1:600), mouse anti-Myospheroid (DSHB CF.6G11, 1:100), rabbit anti-Patj (gift from Dr. Hugo Bellen, 1:500 [34]), and rabbit anti-Yki antibody (gift from Dr. Julia Zeitlinger, 1:1000 [35]). To generate an anti-Patronin antibody, a region 1309–1689 of Patronin isoform I was used to immunize rabbits (ABclonal, USA).
After incubation with primary antibodies, samples were rinsed with washing buffer for 15 min six times and treated with secondary antibodies (Jackson immune research laboratories, 1:200) in washing buffer for 2 h at RT. Samples were rinsed with washing buffer for 10 min six times and PBS for 5 min two times. For DNA staining, samples were treated with 4,6 Diamidine-2-phenylinddedin (DAPI) (Boehringer Mannheim, Germany) at 1:1000 during the last washing in washing buffer. Samples were mounted with vectashield (H-1000, Vector Laboratories, USA). Stained imaginal discs were imaged using a confocal microscope (ZEISS LSM710 or 780). The ZEN program was used for image analysis.
Adult wings were mounted using a wing mounting solution (1:1 mix of Canada balsam-sigma C1795 and methyl salicylate-sigma M6725) and imaged using a light microscope (ZEISS Axio Imager M2) with Axio vision Rel4.8 program. Adult eye pictures were taken using a light microscope (ZEISS KL 1500 LCD Axio cam MRC, Stemi 2000-C) with Axio vision Rel4.8 program. The intensity of immunostaining for PH3 and Dcp-1 was calculated using Image J (NIH, USA). Colocalization was quantified using the Colocalization Finder in Image J. The Manders overlap coefficient was used to quantify the colocalization of immunostaining.
Western blot
Protein samples were separated in SDS-PAGE gel with electrophoresis buffer and transferred to PVDF membrane (Immobilon-P) after 100% methanol activation. Blots were incubated in a blocking solution containing 5% skim milk (232100 from BD Difco) or 3–5% BSA (BSA-BSH from RMBIO) in TBST (10 mM Tris-Cl pH 7.5, 150 mM NaCl, and 0.1% Tween-20) for 30 min at RT. Blots were incubated in blocking solution containing primary antibodies at 4 °C overnight. Primary antibodies were: rabbit anti-Myc (Abcam 9106, 1:5000), mouse anti-β-Actin (Abcam 8224, 1:1000), rabbit anti-Patronin (1:800 [31]), rabbit anti-Yki antiserum (1:4000 [19]), rabbit anti-p-YAP (S127, D9W2I, Cell Signaling Technology 13008, 1:1000), rabbit anti-Klp10A (1:4000 [32]), guinea pig anti-Mer antiserum (1:10,000 [33]), rabbit anti-Histone H3 (Millipore 05–928, 1:10,000), guinea pig anti-Expanded antiserum (gift from Dr. Richard Fehon, 1:5000 [36]), and mouse anti-V5 (Invitrogen 46–0705, 1:5000).
After incubation with primary antibodies, samples were washed for 15 min four times using washing buffer TBST, and incubated with secondary antibodies (Jackson immune research laboratories, 1:10,000) for 1 h at RT, and washed for 15 min four times. Membranes were incubated in ECL solution (Prod #34095, Prod #34080, or Prod #34580, Thermo Fisher Scientific) and exposed to X-ray film. Western blot bands were quantified using Image J (NIH, USA).
S2 cell culture and transfection
S2 cell line (from DGRC) was cultured at 24 °C. Complete medium is composed of serum-free media (Gibco express five SFM, Thermo Fisher Scientific) with 200 mM L-Glutamine and antibiotics (penicillin and streptomycin) as described in the Thermo Fisher Scientific protocol. DNA constructs were purified using a midi-prep kit (Nucleobond Xtra midi, Macherey-Nagel, 740410.50). 1 µg DNA construct was used for transfection using effectene (Qiagen, 301427).
Cultured S2 cells were harvested by centrifugation and washed with PBS (pH 7.4) and centrifuged again. Cell pellets were mixed with lysis buffer (pH 7.5, 20 mM HEPES, 2.5 mM EDTA, 1 mM DTT, 5% glycerol, 100 mM KCl, 0.05% Triton X-100, protease inhibitor cocktail, phosphatase inhibitor, and PMSF) for 30 min at 4 °C. PhosSTOP (Roche, 4906837001) was used as phosphatase inhibitor. The samples were centrifuged at 13,000 rpm for 20 min, and protein concentration of supernatants was measured. Supernatants were mixed with 5x sample buffer and boiled at 94 °C for 10 min and kept at −20 °C.
Transgene construction
A DNA fragment encoding Patronin isoform I from pMT-GFP-Patronin (Addgene #31753) was inserted into pUAST-attB vector to make pUAST-attB-Patronin construct. Using pAC5.1-Mer [37], Mer coding fragment was inserted into pAC5.1-2xMyc vector for making pAC5.1-2x-Myc Mer construct. pUAST-attB-Klp10A construct was made, using pENTR4-Klp10A (gift from Dr. Kim McKim [38]). pAC5.1 His-lacZ (Thermo Fisher Scientific V411020), pAC5.1-ex-V5, pBluescript SK(−), and yki-V5 (gift from Dr. Georg Halder) were used in S2 cell experiments.
PCR was done using Prime star HS DNA polymerase (Takara, R010A) with primers described in Supplementary Table 1. PCR products were separated in 1% agarose gel (Seakem LE agarose for gel electrophoresis, Lonza, 50004), purified using accuprep gel purification kit (Bioneer, Korea), and sequenced (Solgent, Korea).
Generation of transgenic fly lines
cDNA source of pUAST-Patronin line was derived from pMT-GFP-Patronin construct (Addgene #31753). cDNA source of pUAST-Klp10A line was derived from pENTR4-Klp10A construct [38]. Transgenic lines were generated by BestGene (USA) using P[acman] attP strains; PBac{yellow[+]-attP-3B}VK00002 (BDSC 9723, 2nd chromosome) and PBac{yellow[+]-attP-9A}VK00013 (BDSC 9732, 3rd chromosome). pUAST-Patronin (2nd) and pUAST-Klp10A (3rd) are homozygote viable. pUAST-Klp10A (3rd) shows wing blisters by leaky basal expression at 25 °C.
Clonal analysis
Mutant clones were generated by FLP-mediated recombination [39] using the following lines: hs-FLP; FRT42D Ubi-GFPNLS (gift from Dr. Georg Halder), y w ey-FLP, P{5xgIBS-lacZ.38-1}TPN1; FRT42D (BDSC 5616), FRT42D (BDSC 1802), w; FRT42D PatroninEY05252 (modified from Kyoto 114436), and y w; FRT42D Ubi-GFPNLS . For hs-FLP clones, larvae were heat-shocked for 1 h at 37 °C once a day, starting from 2 days after egg laying till late second or early third-instar larval stage. Clones in wing discs were identified by the GFP marker. Clones in adult wings were unmarked. ey-FLP clones in adult eyes were identified by two copies of w+ eye color marker in the PatroninEY05252 allele.
Lethality test
Embryos were collected on grape juice plate with yeast paste at 25 °C to determine the lethal phase of mutants. Homozygote and heterozygote mutant embryos and larvae were distinguished by the absence or presence of the CyO-GFP marker using a fluorescence microscope (Nikon SMZ1500).
Protein extraction from adult flies
Twenty female flies collected in the Eppendorf tube were frozen by liquid nitrogen. Flies were ground using Dounce tissue grinders (Tight, Corning) with lysis buffer (20 mM HEPES, 2.5 mM EDTA, 1 mM DTT, 5% glycerol, 100 mM KCl, 0.05% Triton X-100, protease inhibitor cocktail, PhosSTOP, and PMSF) on ice. Samples were centrifuged at 13,000 rpm for 30 min at 4 °C. After the removal of the lipid layer, protein concentration of the supernatant was measured. Supernatants were mixed with a 5x sample buffer and boiled at 94 °C for 10 min and kept at −20 °C.
Immunoprecipitation assay
S2 cells transfected with 1 µg DNA constructs were harvested by centrifugation and washed with PBS (pH 7.4). Pelleted cells were mixed with lysis buffer (pH 7.5, 20 mM HEPES, 2.5 mM EDTA, 1 mM DTT, 5% glycerol, 100 mM KCl, 0.05% Triton X-100, protease inhibitor cocktail, PMSF, and PhosSTOP). After centrifugation at 13,000 rpm for 20 min, supernatants were collected. For testing the physical interaction of proteins, we used SureBeads™ Protein A/G Magnetic Beads (Bio-Rad).
In the preclearing step, samples were mixed with pre-washed magnetic beads on the rotator at 4 °C for 1 h and loaded in a magnetic rack (Bio-Rad). The supernatant was collected. Simultaneously, primary antibodies were mixed with magnetic beads in lysis buffer and rotated for 1 h at RT. The magnetic bead–antibody mixture was loaded in a magnetic rack, and the supernatant was removed. Primary antibodies were mouse anti-Myc 9E11 (Santa Cruz Sc-47694, 0.2 µg), mouse anti-IgG (Vector I-2000, 0.2 µg), rabbit anti-V5 (Abcam 9116, 2 µg), and rabbit anti-IgG (2 µg).
Precleared samples were added to tubes containing a magnetic bead–antibody mixture, and put on the rotator for 2 h or overnight at 4 °C. Samples were then loaded in a magnetic rack to collect the proteins bound to the magnetic beads-antibody mixture. Samples in the pellet were washed with lysis buffer 5–7 times, mixed with 1x sample buffer, and boiled at 94 °C for 10 min. Samples were loaded in a magnetic rack to remove magnetic beads. The supernatant containing immunoprecipitated proteins was transferred into a new tube and kept at −20 °C.
Generation of P-element excision lines
Patronin revertants were generated by precise excision of the P{EPgy2} element from PatroninEY05252. PatroninEY05252/CyO (BDSC 16647) was mated with Sp/CyO; P{Δ2–3}99B/TM6B. In the F1 step, PatroninEY05252/CyO; P{Δ2–3}99B/+ single males were mated with In(2LR)Gla Bc/CyO virgins. In the F2 step, single white eye males with CyO were collected to establish balanced lines. We confirmed that Patroninrev homozygote flies are viable.
Synthesis of double-strand RNA
cDNAs for Patronin, Klp10A, ex, yki, Mer, and pBluescript SK(−) were used as templates to generate double-strand RNA (dsRNA). Primers were synthesized (Bioneer, Korea) (Supplementary Table 1). Primer target sites for Patronin dsRNA, Klp10A dsRNA, ex dsRNA, and control dsRNA were selected as described [31, 40,41,42]. Target sites for yki dsRNA and Mer dsRNA were selected using the SnapDragon from Harvard medical school. Using the dsRNA template product, in vitro transcription was carried out using EZ™ T7 High Yield In Vitro Transcription Kit (Enzynomics, EZ027S) at 37 °C overnight. Next, the RNA mixture was treated with DNase Ι (Amplification grade, Thermo Fisher Scientific, 18068015) for 30 min at 37 °C. Samples were purified using phenol–chloroform isoamyl alcohol mixture (Sigma, 77619), and kept at −80 °C. 30 µg dsRNA was transfected into S2 cells and incubated at 24 °C. dsRNA transfection was performed using the protocol of the Drosophila RNAi screening center at Harvard medical school.
Quantification and statistical analysis
Wing and eye sizes were measured using the Image J program (NIH, USA). Wing and eye images were taken from representative samples with average size. Statistical analysis of organ sizes was performed using Prism 8 (GraphPad, USA). Sample mean (x̅) and sample standard deviation (s) were used when samples were selected from a population. Population mean (m) and population standard deviation (σ) were used when samples were not selected. We used unpaired two-tailed Student's t test with standard deviation (SD). If the SD of the two groups were significantly different based on F-test, we used Welch’s correction. Significance was defined by the rule of GraphPad program, not significant ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Knockdown of Patronin reduces organ size
To test whether Patronin is involved in organ development, we performed a tissue-specific knockdown of Patronin using the GAL4-UAS system [43]. We used three independent UAS-RNAi lines targeting different exons of the Patronin gene (Fig. 1a). Patronin knockdown at RT using RNAiGD11946 by engrailed (en)-GAL4 (hereafter labeled as en > Patronin RNAi) resulted in a wing reduction with blisters in the posterior region (Fig. 1c). Patronin RNAiGD11946 with patched (ptc)-GAL4 led to a decrease in the targeted area between the veins L3 and L4 along the anterior–posterior (AP) boundary (Figs. 1d–f′ and S1a–d′), often causing blisters or small wing notching (Fig. S1c). Patronin RNAiGD11946 using nubbin (nub)-GAL4, which drives GAL4 expression in the wing pouch, resulted in reduced wings with variable sizes of blisters (Fig. 1g, h). In addition, most wings with Patronin RNAiGD11946 were significantly reduced in size (32.2 ± 5% reduction) (Fig. S1m, o, q).
a Schematic diagram of the coding region of the Patronin gene. Three Patronin RNAi (Patr i) targets are indicated in blue letters. This diagram is based on the information of InterProScan (EMBL-EBI, UK) and FlyBase (USA). CH (Calponin Homology domain), CC (Coiled-Coil domain), and CKK domain. b, c Knockdown of Patronin results in smaller wings with blisters at RT. b en > GFP/+ and c en > GFP > Patronin RNAiGD11946 (N = 24; 41.6%). Red arrows in (c, e, h) indicate blisters in the wing. N is the total number of flies scored for wing blisters. Percentage numbers in (c, e, h) indicate % flies showing blisters. Scale bars in (b, d, d′, g) are 300 µm. d, e Patronin RNAi by ptc-GAL4 reduces wing size. d ptc > GFP/+ (n = 7, x̅ ± s = 1 ± 0.022) and e ptc > GFP > Patronin RNAiGD11946 (N = 14; 50%, n = 7, x̅ ± s = 0.851 ± 0.011). n is the number of wings used to measure wing size and the width between L3 and L4 veins. x̅ is relative average (sample mean) wing size and the width between L3 and L4 veins. s is the sample standard deviation. “%,” N, n, x̅, and s are similarly used in all Figures. The dotted line in (e) indicates wing size of control wild-type shown in (d). L2–L5 in (d) indicates the longitudinal veins 2–5. f Quantification of relative wing sizes in (d, e). Unpaired two-tailed t-test. ****p < 0.0001. Error bars are standard deviation. d′, e′ Knockdown of Patronin reduces the width between L3 and L4 veins. d′ Enlarged view of the boxed area in (d) (n = 7, x̅ ± s = 1 ± 0.024). e′ Enlarged view of the boxed area in (e) (N = 14; 50%, n = 7, x̅ ± s = 0.815 ± 0.037). Red dotted lines indicate the width between L3 and L4 veins in the control line in (d′). f′ Quantification of relative L3–L4 width in (d′, e′). g, h Knockdown of Patronin results in smaller wings with blisters. g nub-GAL4/+ and h nub > Patronin RNAiGD11946 (N = 18; 94.4%).
Two other Patronin RNAi lines (RNAiKK100898 and RNAiHMS01547) showed similar blisters when knockdown was induced by en-GAL4 or nub-GAL4 (Fig. S1f, g, i, k). Patronin RNAiKK100898 has an off-target to the bnl gene, but bnl RNAi has no effect in the wing (Fig. S2h–m, o, p). Penetrance of the blister phenotype by RNAiHMS01547 was lower than those of RNAiGD11946 or RNAiKK100898 (Figs. 1 and S1). However, Patronin knockdown by RNAiHMS01547 using nub-GAL4 caused strong wing reduction in 50% of wings examined (N = 80) (Fig. S1l). Similar wing phenotypes by three independent RNAi lines suggest that RNAi phenotypes are specific.
Furthermore, we generated a UAS-Patronin line for overexpression in the targeted regions of the wing disc (Fig. S1r–s″). Overexpression of Patronin rescued the Patronin RNAi wing phenotypes (Fig. S1o–q). This rescue was unlikely due to titration of GAL4 by an extra copy of UAS, since two independent UAS-GFP lines could not rescue the Patronin RNAi phenotypes (Fig. S2a–g, n).
Patronin mutation affects the size of wing and eye
We examined whether Patronin RNAi affects other organs. Patronin knockdown in the eye disc using eyeless (ey)-GAL4 showed no consistent eye abnormalities. Hence we utilized Dicer2 (Dcr2) to enhance the RNAi efficiency [44]. Patronin RNAiGD11946 with Dcr2 overexpression showed no eye size reduction at 25 °C (Fig. S3a, b, g) but caused a consistent reduction at 29 °C (Fig. S3d, e, h). In contrast, Patronin RNAiKK100898 resulted in stronger eye reduction even at 25 °C (Fig. S3a, c, d, f–h).
To further confirm the Patronin RNAi effects, we examined Patronin mutants. PatroninEY05252 and Patronink07433 (Fig. S4a) are homozygous embryonic lethal. These P-element insertion mutants failed to complement the deficiency Df(2R)BSC355 (54B16-54C3) uncovering the Patronin gene. Ubiquitous Patronin overexpression by Ubiquitin 63E promoter rescued the lethality of PatroninEY05252 but not Patronink07433. Hence, PatroninEY05252 lethality was due to the P-element insertion, consistent with the result that P-element mobilization reverted the lethality (Fig. S4b–e). In contrast, Patronink07433 may have a second site lethal mutation. PatroninEY05252 has been described as a hypomorphic mutant [12]. Patronin levels were reduced in PatroninEY05252/+ heterozygote in western blot (Fig. S4f). Embryonic lethality of Patronin mutations indicates that Patronin is essential for early development. Similarly, the ubiquitous knockdown of Patronin by tubulin (tub)-GAL4 also caused lethality.
Since PatroninEY05252 mutants are lethal, we generated PatroninEY05252 mutant clones using the FLP-FRT method [39]. Patronin protein was strongly decreased or undetectable in PatroninEY05252 mutant clones in wing discs (Fig. S4g–g″). Adult wings with unmarked mutant clones showed size reduction and blisters (Fig. S4h–j), as Patronin RNAi wings. Eyes containing PatroninEY05252 mutant clones also showed about 16.9 ± 6.4% eye reduction, compared with the eyes containing wild-type control clones (Fig. S4k–m).
Knockdown of Patronin leads to activation of caspase
Cell survival and proliferation contribute to organ size. Thus, we examined Patronin RNAi effects on cell death. Flies were cultured at 29 °C to increase the RNAi effects on cell death. Normal wing disc showed even distribution of DAPI stain in both anterior and posterior compartments (Fig. 2a′, c′). In contrast, en > Patronin RNAiHMS01547 (Fig. 2b′) or en > Patronin RNAiGD11946 (Fig. 2d′) showed partial loss of DAPI stain in the posterior region. We tested whether such defects are related to apoptosis by staining for Dcp-1. The results showed ectopic cleaved Dcp-1 stain in the posterior domain (Fig. 2f′, h′), suggesting that loss of Patronin causes apoptosis. In accordance with the low penetrance of the adult wing phenotype of Patronin RNAiHMS01547 (Fig. S1f), this RNAi line showed relatively strong Dcp-1 stain with a low frequency (N = 13; 23%). On the contrary, Dcp-1 stain in Patronin RNAiGD11946 wing discs was weak (Fig. 2h′), but its penetrance was high (N = 6; 100%), consistent with the high penetrance of its wing phenotype (Fig. S1o). Since Patronin RNAiGD11946 showed high penetrance of the larval and adult wing phenotypes, we used this RNAi line for further analysis.
a–a″ en > GFP/+ control wing disc. a GFP, a′ DAPI, a″ Merge. “A” and “P” indicate anterior and posterior, respectively. Scale bars in a–h are 50 µm. b–b″ en > GFP > Patronin RNAiHMS01547 shows partial loss of DAPI stain in the posterior region (arrow) (N = 5; 100%). b GFP, b′ DAPI, b″ Merge. Dotted lines in (a′–h′) indicate the anterior–posterior (AP) boundary. N is the total number of wing discs scored for the DAPI stain phenotype. Percentage numbers in (b′, d′, f′, h′) indicate % wing discs that show partial loss of DAPI stain or increased Dcp-1 stain. c–c″ en > GFP > Dcr2 shows a normal DAPI pattern in the posterior region. c GFP, c′ DAPI, c″ Merge. d–d″ en > Patronin RNAiGD11946 > GFP > Dcr2 shows partial loss of DAPI stain in the posterior region (arrow) (N = 6; 100%). d GFP, d′ DAPI, d″ Merge. e–e″ en > GFP/+ control wing disc shows no cleaved Dcp-1 stain. e GFP, e′ Dcp-1, e″ Merge. f–f″ en > GFP > Patronin RNAiHMS01547 shows ectopic Dcp-1 stain in the posterior region (arrow) (N = 13; 23.1%). f GFP, f′ Dcp-1, f″ Merge. g–g″ en > GFP > Dcr2 control wing disc. g GFP, g′ Dcp-1, g″ Merge. h–h″ en > Patronin RNAiGD11946 > GFP > Dcr2 shows Dcp-1 puncta stain in the posterior region (N = 6; 100%). h GFP, h′ Dcp-1, h″ Merge. White dotted circles indicate the regions of Dcp-1 puncta stain. All tests in a–h″ were performed at 29 °C.
Next, we examined whether Patronin RNAi might affect cell size by examining the adherens junction marker Arm. In wing discs with en > Patronin RNAiGD11946, there was no obvious cell size difference between anterior and posterior compartments (Fig. S5a–d″). We also tested whether the loss of Patronin influences cell proliferation. P/A ratios in size and PH3 density were similar in control and Patronin RNAi disc (Fig. S5e–h). These data suggest that loss of Patronin affects cell survival but not cell size and proliferation in the larval wing disc.
Patronin RNAi phenotypes are suppressed by reducing microtubule minus-end destabilizing factors
Since Patronin is required for microtubule protection, we tested whether reduced wing size by Patronin RNAi can be suppressed by depleting microtubule severing factors. Spastin (Spas) is a severing enzyme that destabilizes microtubule at the minus end, whereas Katanin 60 (Kat60) is a plus-end severing depolymerase [45]. The knockdown of Spas or Kat60 by en-GAL4 caused no significant wing size defect (Fig. 3a–c, g, h). Kat60 RNAi lines could not suppress Patronin RNAi phenotypes (Fig. 3i–k). In contrast, spas RNAi lines strongly suppressed Patronin RNAi phenotypes (Fig. 3i, o, p). Hence, Patronin antagonizes the Spas function at the minus end but has little effect on the Kat60 function at the plus end. Eb1 is a microtubule growth regulator at the plus end [45]. Overexpression of Eb1 did not significantly affect wing development in wild-type condition (Fig. 3a, d) and could not suppress the Patronin RNAi phenotype (Fig. 3i, l).
a–h Effects of Kat60 RNAi, Klp10A RNAi, spas RNAi, or Eb1 overexpression in wild-type wing. All show normal wings. a en > GFP/+, b en > GFP > Kat60 RNAiKK108203, c en > GFP > Kat60 RNAiJF03012, d en > GFP > Eb1-EGFP, e en > GFP > Klp10A RNAiGD8358, f en > GFP > Klp10A RNAiHMS00920, g en > GFP > spas RNAiJF02724, h en > GFP > spas RNAiHMC03560. Scale bars in (a, l, q) are 300 µm. i–p Rescue of Patronin RNAi phenotype by Klp10A RNAi or spas RNAi, but not by Kat60 RNAi or Eb1 overexpression. i en > Patronin RNAiGD11946/+ (N = 20; 70%), j en > Patronin RNAiGD11946 > Kat60 RNAiKK108203 (N > 10; 90–100%), k en > Patronin RNAiGD11946 > Kat60 RNAiJF03012 (N > 10; 90–100%), l en > Patronin RNAiGD11946 > Eb1-EGFP (N = 24; 100%), m en > Patronin RNAiGD11946 > Klp10A RNAiGD8358 (N > 10; 0%), n en > Patronin RNAiGD11946 > Klp10A RNAiHMS00920 (N > 10; 0%), o en > Patronin RNAiGD11946 > spas RNAiJF02724 (N = 10; 20%), p en > Patronin RNAiGD11946 > spas RNAiHMC03560 (N = 12; 25%). Red arrows in (i–l, r) indicate wing blisters. q–s Klp10A RNAi can rescue Patronin RNAi phenotypes. q en > GFP/+ (n = 7, x̅ ± s = 1 ± 0.018), r en > Patronin RNAiGD11946/+ (N = 25; 80%, n = 10, x̅ ± s = 0.748 ± 0.064), s en > Patronin RNAiGD11946 > Klp10A RNAiGD8358 (N = 18; 0%, n = 8, x̅ ± s = 0.867 ± 0.028). t Quantification of relative wing sizes in (q–s) with unpaired two-tailed t-test. ***p < 0.001, ****p < 0.0001. Error bars are standard deviation.
Next, we examined the relationship between Patronin and Klp10A, based on their antagonistic relationship in mitotic spindles [7, 11]. We used two independent Klp10A RNAi lines to test the antagonistic interaction between Patronin and Klp10A. Loss of Klp10A causes lethality [38], but Klp10A RNAi by en-GAL4 did not noticeably affect wing size at 25 °C (Fig. 3a, e, f). Under this condition, Klp10A knockdown rescued Patronin RNAi phenotypes (Figs. 3i, m, n, r–t and S6b–d), suggesting that Patronin acts antagonistically to Klp10A in wing development.
Patronin RNAi wing phenotypes are suppressed by Diap1 overexpression or reduction of Hippo signaling factors
Because Patronin RNAi induces ectopic activation of Dcp-1(Fig. 2f′, h′), we tested whether Diap1 overexpression can suppress wing phenotypes of Patronin RNAi. Diap1 overexpression did not affect the wing size in the wild-type background (Fig. 4a, b, g), but rescued Patronin knockdown phenotypes (Figs. 4d, e, g and S6g–i).
a–f Patronin RNAi phenotype is rescued by overexpression of Diap1 but not CycE. a en > GFP/+ (n = 7, x̅ ± s = 1 ± 0.032), b en > GFP > Diap1 (n = 7, x̅ ± s = 0.995 ± 0.042), c en > GFP > CycE (n = 7, x̅ ± s = 1.036 ± 0.064), d en > Patronin RNAiGD11946/+ (N = 34; 67.6%, n = 7, x̅ ± s = 0.687 ± 0.074), e en > Patronin RNAiGD11946 > Diap1 (N = 20; 20%, n = 11, x̅ ± s = 0.921 ± 0.096), and f en > Patronin RNAiGD11946 > CycE (75%, n = 6, x̅ ± s = 0.710 ± 0.064). Red arrows in (d, f, l, s) indicate wing blisters. Scale bars in (a, h, k, q) are 300 µm. g Quantification of relative wing sizes in (a–f). h–j wts/+ and hpo/+ heterozygote mutant wings. h w1118 (n = 6, x̅ ± s = 1 ± 0.038), i wtsP2/+ (n = 6, x̅ ± s = 1.088 ± 0.041), j hpoKS240/+ (n = 7, x̅ ± s = 0.8998 ± 0.043). k–n Reduction of Wts or Hpo function can rescue the Patronin RNAi phenotypes. k en > GFP/+ (n = 7, x̅ ± s = 1 ± 0.018), l en > Patronin RNAiGD11946/+ (N = 25; 80%, n = 10, x̅ ± s = 0.748 ± 0.064), m en > Patronin RNAiGD11946; wtsP2/+ (N = 32; 59.3%, n = 7, x̅ ± s = 1.023 ± 0.025), n en > Patronin RNAiGD11946/hpoKS240 (N = 14; 7.1%, n = 7, x̅ ± s = 0.9135 ± 0.061). Dotted lines in (m, n) indicate the wing size of en > Patronin RNAiGD11946/+ shown in (l). Experiments for this figure h–p and Fig. 3q–t were performed simultaneously. Wings in (k, l) were shown in Fig. 3q, r, respectively. o Quantification of relative wing sizes in (h–j). p Quantification of relative wing sizes in (k–n). q–t Overexpression of Yki rescues Patronin RNAi phenotypes. q en > GFP/+ (n = 7, x̅ ± s = 1 ± 0.049), r en > GFP > Yki-V5 (n = 6, x̅ ± s = 1.117 ± 0.046), s en > Patronin RNAiGD11946/+ (N = 20; 70%, n = 5, x̅ ± s = 0.606 ± 0.088), and t en > Patronin RNAiGD11946 > Yki-V5 (N = 31; 38.7%, n = 7, x̅ ± s = 1.106 ± 0.013). Dotted line in (r) indicates the control wing size shown in (q). Experiments in this figure (q–u) and Fig. 3a–p were performed simultaneously. Wings in (q, s) were shown in Fig. 3a, i, respectively. u Quantification of relative wing sizes in (q–t). Statistical analysis in (g, o, p, u) by unpaired two-tailed t-test. ns: p > 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Error bars are standard deviation.
In addition, we examined the effects of another cell death inhibitor p35. Unexpectedly, control wings overexpressing p35 by en-GAL4 showed a mild wing reduction (12.7 ± 4.2%) (Fig. S6j, l, o). When p35 was overexpressed with Patronin RNAi, the wing size was rescued in ~46% of wings examined (Fig. S6k, m, o). In contrast, overexpression of CycE failed to suppress Patronin RNAi phenotypes (Fig. 4d, f, g). Hence, reduced wing size by Patronin RNAi is mainly due to abnormal cell death.
Since Diap1 expression is activated by Yki [17], we checked the effects of reducing Hpo or Wts that antagonize the Yki function. Patronin RNAi effects on wing size were considerably restored in hpoks240/+ or wtsP2/+ conditions, respectively (Fig. 4l–n, p). Furthermore, wtsP2/+ mutation reduced the Dcp-1 level in the posterior region of en > Patronin RNAi wing disc (Fig. S7d′, e′), implying that Patronin is functionally related to Hippo signaling.
Furthermore, Yki overexpression by en-GAL4 suppressed the Patronin RNAi wing phenotypes (Fig. 4s–u), whereas it weakly increased the wild-type wing size approximately by 11.7 ± 4.6% (Fig. 4q, r, u). We checked whether Patronin RNAi may alter Yki levels in wing discs. Although there was a weak reduction of Yki levels in the posterior domain of Patronin RNAi wing discs, a similar posterior Yki reduction was also seen in control wing discs (Fig. S8a′, b′), suggesting that Patronin RNAi does not decrease Yki levels in the wing disc. Similarly, Patronin RNAi did not reduce the level of Diap1-lacZ (Fig. S8c′, d′). Alternatively, we performed western blot experiments. In PatroninEY05252/+ adult flies, Yki and p-Yki levels were decreased to 62.5 ± 23.3% and 30.0 ± 0.2%, respectively (Fig. S8e–g, [46]). Hence, reduced Patronin function leads to Yki levels reduction in adult tissues, although such change was not clearly detected in wing discs.
Patronin is enriched at subapical cell junctions and its loss impairs cell organization
To further characterize the role of Patronin in Hippo signaling, we examined the relationship between Patronin and Mer, a microtubule-associated protein [26,27,28,29] involved in the Hippo pathway. We checked Patronin localization in wing disc epithelia, using a Patronin:YFP knock-in fly line [12]. Patronin:YFP was expressed in all wing disc cells, but a higher level was detected in two stripes of cells that flank the dorsoventral boundary. This stripe pattern was observed mostly in the anterior compartment (Fig. 5a, c). In these cells, Patronin-YFP was found mainly in the medial region at the apical level (Fig. 5b), while it was enriched in the cell membrane at the subapical level (Fig. 5d). The stripe pattern of Mer was also observed mostly in the anterior compartment (Fig. 5a′, c′). Mer was enriched at the apical and subapical regions (Fig. 5b′, d′). Patronin-YFP and Mer showed significant colocalization along cell membranes at the subapical level (Fig. 5d–d″) which can also be seen in z-stack images (Fig. 5e–e″).
a–a″ Localization of Patronin:YFP and Mer at the apical section of dorsoventral (DV) boundary region of the wild-type wing disc. Patronin:YFP is stained with an anti-GFP antibody. a GFP, a′ Mer, a″ Merge. Scale bars in (a–e) are 10 µm. Patronin is enriched in the medial region within Mer-enriched cell boundaries. Yellow arrows in (a, a′, c, c′) denote the stripe pattern in the anterior compartment. White arrows in (a–c″) indicate the AP boundary. b–b″ Magnification of yellow boxed area shown in (a). b GFP, b′ Mer, b″ Merge. c–c″ Localization of Patronin:YFP and Mer at the subapical section of wild-type wing disc. c GFP, c′ Mer, c″ Merge. d–d″ Magnification of yellow boxed area shown in (c). d GFP, d′ Mer, d″ Merge. Patronin and Mer show overlapping localization at the cell boundaries with Mander’s overlap coefficient 0.924. e–e″ Z-stack images of wing disc shown in (a–d″). e GFP, e′ Mer, e″ Merge. f–g‴ Z-stack images of two wing disc samples of en > Patr iGD11946 > GFP > Dcr2 at 29 °C. f, g GFP, f′, g′ Patj, f″, g″ Myospheroid (Mys), f‴, g‴ Merge. Patronin knockdown in the posterior (P) region causes sinking of the apical region marked by Patj stain (arrows in (f′, g′)) (N = 6; 83.3%). Mys stain shows a relatively smooth basal surface. Dotted lines in (f′–g″) indicate the AP boundary. N is the total number of wing discs scored for apical defects. Percentage numbers in (f–g‴) indicate % wing discs showing the abnormal apical region. Scale bars in (f, g) are 50 µm.
Next, we tested whether Patronin knockdown affects Mer. Cross section of en > Patronin RNAiHMS01547 wing discs showed that the apical surface of the posterior region was slightly tilted down (Fig. S9c–c″). A wing disc consists of the peripodial epithelium and the underlying disc proper epithelium. Serial sections showed that the junctional Mer of the posterior cells are located more basally than the anterior cells (Fig. S9d, d′). For example, the anterior region of Section 1 showed both peripodial cells and small disc proper cells, whereas the posterior region showed only large peripodial cells. Disc proper cells in the posterior region began to appear in the more basal Section 2, indicating that junctional Mer in the posterior domain is located basal to that in the anterior domain. We also checked whether Patronin RNAiGD11946 shows similar changes in the apical marker Patj. en > RNAiGD11946 wing discs showed apical sinking in local areas of the en domain (Fig. 5f–g‴), indicating that the loss of Patronin impairs apical cell organization.
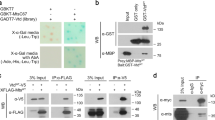
Patronin physically and genetically interacts with Mer and Ex
We also examined whether Patronin and Mer are physically associated. Endogenous Patronin and Myc-Mer were co-immunoprecipitated in S2 cells (Fig. 6a–c), consistent with their colocalization (Fig. 5d–d″). We then tested whether Patronin genetically interacts with Mer. Mer loss-of-function did not induce obvious wing overgrowth [47], as Mer and Ex have redundant functions [36, 47, 48]. Mer RNAi itself showed no significant effect (Fig. 6d, e, i) or a marginal increase (~12.8 ± 3.5%) in wing size (Fig. S10a, b, e). Two Mer RNAi lines similarly suppressed Patronin RNAi wing phenotypes (Figs. 6f, g, i and S10c–e).
a Immunoprecipitation between Patronin and 2xMyc-Mer. A representative of three assays is shown. Arrows in (a) indicate the 2xMyc-Merlin bands. Patronin and Myc-Mer proteins migrate slightly slower in immunoprecipitated samples than in the input mixture. b Mer protein level is decreased by Mer dsRNA in S2 cells. Arrows in (b) denote the Merlin bands. c 2xMyc-Mer protein level is increased in Mer overexpressed S2 cells. Arrow in (c) denotes the 2xMyc-Merlin band. d–h Knockdown of Mer or Ex can rescue Patronin RNAi phenotypes. d en > GFP/+ (n = 7, x̅ ± s = 1 ± 0.018), e en > GFP > Mer RNAiGD1484 (n = 8, x̅ ± s = 0.972 ± 0.030), f en > Patronin RNAiGD11946/+ (N = 25; 80%, n = 10, x̅ ± s = 0.748 ± 0.064), g en > Patronin RNAiGD11946 > Mer RNAiGD1484 (N = 15; 6.6%, n = 7, x̅ ± s = 0.961 ± 0.031), h en > Patronin RNAiGD11946/exe1 (N = 14; 7.1%, n = 7, x̅ ± s = 1.040 ± 0.013). Red arrow in (f) indicates wing blisters. Dotted lines in (g, h) indicate the wing size of en > Patronin RNAiGD11946/+ shown in (f). Scale bar in (d) is 300 µm. Experiments for this figure (d–i) and Figs. 3q–t and 4h–p were performed simultaneously. Wings in (d, f) were shown in Figs. 3q, r and 4k, l, respectively. i Quantification of relative wing sizes in (d–h) with unpaired two-tailed t-test. ns: p > 0.05 and ****p < 0.0001. Error bars are standard deviation. j Immunoprecipitation assay between Patronin and Ex-V5. A representative of three assays is shown. Arrows in (j) show the Ex-V5 and endogenous Merlin bands. k Levels of Ex protein bands are decreased by ex dsRNA in S2 cells.
Since Mer and Ex function together, we checked whether Patronin also interacts with Ex. Ex-V5 co-immunoprecipitated endogenous Patronin and Mer in S2 cells (Fig. 6j, k), indicating their association. We also checked the genetic interaction between Patronin and ex, using three ex mutant alleles. Wings of heterozygotes for these alleles were slightly larger or smaller than that of control (Fig. S10f–j), but rescued Patronin RNAi wing phenotypes (Figs. 6f, h, i and S10l–o). Taken together, Patronin is physically associated with Mer and Ex, and Patronin loss phenotypes can be restored by reducing Mer or Ex.
Klp10A overexpression activates cell death and reduces organ size
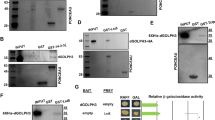
To further test the antagonistic relationship between Patronin and Klp10A (Fig. 3), we examined whether Klp10A overexpression causes similar wing defects seen by Patronin RNAi. Klp10A overexpression led to a size reduction with wing blisters and occasional notching (Figs. 7a–c and S11a, b). Klp10A overexpression using ey-GAL4 also decreased the eye size (Figs. 7d–f and S11c–e). The phenotypes of Klp10A overexpression were similar or more severe than those of Patronin RNAi. We tested whether Patronin RNAi can affect Klp10A levels. When Patronin was knocked down in S2 cells, Klp10A levels were increased by about 2.5 ± 0.5-fold when normalized to the actin level (Figs. 7g and S11f, g). Furthermore, Klp10A overexpression by en-GAL4 resulted in a significant reduction of the Patronin level in the posterior region (Figs. 7h′ and S11h′), supporting their antagonistic relationship.
a, b Overexpression of Klp10A reduces wing size. a en > GFP/+ (n = 10, x̅ ± s = 1 ± 0.015) and b en > GFP > Klp10A (N = 34; 61.7%, n = 11, x̅ ± s = 0.524 ± 0.019). Scale bar in (a) is 300 µm. Red arrow in (b) indicates wing blisters. c Quantification of relative wing sizes in (a, b). Statistical analysis in (c, f) by unpaired two-tailed t-test. ****p < 0.0001. Error bars are standard deviation. d, e Overexpression of Klp10A reduces eye size at 29 °C. d ey-GAL4/+ (n = 11, x̅ ± s = 1 ± 0.059), e ey > Klp10A (N = 44; 100%, n = 10, x̅ ± s = 0.640 ± 0.046). Scale bar in (d) is 150 µm. x̅ is relative eye average size in (d, e). f Quantification of relative eye size in (d, e). g Klp10A level is increased in Patronin-depleted S2 cells (N = 3, m ± σ = 2.520 ± 0.496). m is average (population means) of Klp10A band intensity relative to actin. σ is the population standard deviation. h–h″ en > GFP > Klp10A decreases Patronin level (N = 6; 100%). h GFP, h′ Patronin, h″ Merge. Dotted lines in (h′–k′, m′) indicate the AP boundary. The yellow arrow in h′ denotes reduced Patronin level in the posterior region. Scale bars in (h–m) are 50 µm. i–i″ en > GFP > Klp10A decreases DAPI level in the posterior region (arrow) (N = 8; 100%). i GFP, i′ DAPI, i″ Merge. j–j″ en > GFP > Klp10A increases Dcp-1 level in the posterior region (arrow) (N = 5; 100%). j GFP, j′ Dcp-1, j″ Merge. k–k″ en > GFP > Klp10A increases Mer and Arm level at the periphery of a defective islet structure (yellow arrows) (N = 6; 83.3%). k Mer, k′ Arm, k″ Merge of Mer, Arm, and GFP. l–l″ Z-stack images of the wing disc shown in (k–k″). l Mer, l′ Arm, l″ Merge of Mer, Arm, and GFP. Red arrows in l–l″ indicate a sinked apical region marked by Mer and Arm stain (N = 6; 100%), with a relatively flat basal surface. White arrows in l–l″ indicate the AP boundary. m–m″ en > GFP > Klp10A decreases Yki level (arrow) (N = 3; 100%). m GFP, m′ Yki, m″ Merge.
Next, we examined whether Klp10A overexpression also induces apoptosis. Klp10A overexpression led to a decreased DAPI stain in the posterior region (Figs. 7i′ and S12e′). Klp10A overexpression also increased the Dcp-1 level in the posterior region of all wing discs examined (Fig. 7j′). The increase of Dcp-1 levels was also seen in Patronin RNAi wing discs (Fig. 2f′, h′). Thus, Klp10A overexpression induces apoptosis, resulting in reduced and deformed wings. Wing discs with Klp10A overexpression showed apically compressed islets of cells surrounded by high levels of Mer and Arm (Figs. 7k–l′ and S12f, f′), suggesting that Klp10A overexpression also causes defects in cell organization. In addition, patches of Klp10A overexpressing cells showed reduced Yki levels (Figs. 7m′ and S12g′).
Suppression of Klp10A gain-of-function phenotypes by p35 but not Diap1
We noted that Klp10A overexpression results in a reduction of Patronin and Yki levels (Fig. 7). Because Klp10A overexpression causes cell death, the reduction of these proteins might be due to cell death. However, Patronin levels were not noticeably recovered by either Diap1 or p35 (Fig. S13b′, d′, f′). Yki reduction by Klp10 overexpression was also not recovered by p35 (Fig. S13h′, l′). In the case of Diap1 overexpression, Yki levels were partially recovered (Fig. S13h′, j′). Thus, the reduction of Patronin levels is not simply due to cell death, while Yki reduction by Klp10A overexpression may be partially due to cell death (Fig. S13h′, j′, l′).
Next, we tested whether ectopic cell death is responsible for the wing size reduction by Klp10A overexpression. Diap1 overexpression led to partial (68.2% of wings examined) or full (31.8%) wing suppression (Fig. 8b, d–f). p35 overexpression led to partial (93.1%) or full (6.9%) wing suppression (Fig. 8h, j, k). Hence, cell death is a significant cause of the Klp10A wing phenotypes. We also tested whether Klp10A wing phenotypes can be suppressed by overexpressing Yki. However, Yki overexpression in en > Klp10A flies caused ~76.5% (N = 34) pupal lethality, while overexpression of Klp10A alone resulted in about 35.9% (N = 39). To avoid such lethal effect of Yki overexpression, we used wtsP2/+ mutation and found that wtsP2/+ mutation partially suppresses the wing phenotype of Klp10A overexpression (Fig. 8m–o) and decreases pupa lethality to ~7.9% (N = 38), which supports genetic interaction between Klp10A and Hippo signaling.
a–e Overexpression of Diap1 partially rescues the Klp10A overexpression phenotypes at RT. a en > GFP/+ (n = 14, x̅ ± s = 1 ± 0.040), b en > GFP > Klp10A/+ (N = 19; 94.7%, n = 15, x̅ ± s = 0.410 ± 0.151), c en > GFP > Diap1 (n = 13, x̅ ± s = 0.977 ± 0.018), d en > GFP > Klp10A > Diap1 (N = 44; 68.2%, n = 13, x̅ ± s = 0.942 ± 0.023), and e en > GFP > Klp10A > Diap1 (n = 20, x̅ ± s = 0.588 ± 0.082). Red arrows in (b, e, h, j, m, n) indicate wing blisters. Scale bars in (a, g, l) are 300 µm. f Quantification of relative wing sizes in (a–e). Statistical analysis in (f, k, o) by unpaired two-tailed t-test. ns: p > 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Error bars are standard deviation. g–j Overexpression of p35 partially rescues the Klp10A overexpression phenotype at RT. g en > GFP/+ (n = 15, x̅ ± s = 1 ± 0.037), h en > GFP > Klp10A/+ (N = 38; 97.4%, n = 13, x̅ ± s = 0.583 ± 0.158), i en > GFP > p35 (n = 14, x̅ ± s = 0.895 ± 0.022), j en > GFP > Klp10A > p35 (N = 29; 93.1%, n = 16, x̅ ± s = 0.734 ± 0.064). k Quantification of relative wing sizes in (g–j). l–n Reduction of Wts function partially rescues the Klp10A overexpression phenotype at RT. l en > GFP/+ (n = 12, x̅ ± s = 1 ± 0.028), m en > GFP > Klp10A/+ (N = 25; 100%, n = 14, x̅ ± s = 0.517 ± 0.063), n en > GFP > Klp10A/wtsP2 (N = 35; 94.3%, n = 12, x̅ ± s = 0.656 ± 0.122). o Quantification of relative wing sizes in (l–n). p, p′ en > GFP/+ control wing disc. p Dcp-1 and p′ Merge with GFP. Dotted lines in (p–r) indicate the AP boundary. N = 4, m ± σ = 0.9854 ± 0.040. m is the average (population mean) of the P/A ratio in Dcp-1 stain intensity. σ is the population standard deviation. Scale bars in (p–r) are 50 µm. q, q′ en > GFP > Klp10A/+ shows ectopic Dcp-1 stain in the posterior region (arrow) (N = 6; 100%). q Dcp-1 and q′ Merge with GFP. N = 6, m ± σ = 1.823 ± 0.237. r, r′ en > GFP > Klp10A/wtsP2 shows weaker Dcp-1 stain in the posterior region (arrow) (N = 5; 100%). r Dcp-1 and r′ Merge with GFP. N = 5, m ± σ = 1.582 ± 0.077.
We checked whether the cell death inhibition occurs in larval wing discs. The results showed that high levels of Dcp-1 induced by Klp10A were considerably suppressed by p35 overexpression (Fig. S14b′, d′). However, Diap1 overexpression did not noticeably decrease the Dcp-1 level in wing discs (Fig. S14b′, f′). Because Diap1 can suppress adult wing phenotypes, cell death inhibition by Diap1 may occur during pupal stage, resulting in suppressed adult wings. We then examined the effects of Hippo signaling. Dcp-1 levels were not significantly reduced by Yki overexpression (Fig. S14b′, h′) but suppressed by wtsP2/+ heterozygote condition (Fig. 8p–r′)
Discussion
Role of Patronin and Klp10A in epithelial integrity and cell death
We have shown new roles of Patronin in the control of organ size. Patronin RNAi causes caspase activation and loss of nuclei in the wing disc. The knockdown of Patronin also results in apical compression of the wing epithelium (Figs. 5f–g‴ and S9c–d′), implying a role of Patronin for epithelial integrity. Klp10A overexpression also causes apical compressions in the wing disc, often resulting in the formation of islet-like structures (Fig. 7k–l′). Wing disc cells have both longitudinal microtubules along the apical–basal axis and the circumferential microtubules adjacent to the adherens junction [49]. Loss of Patronin or Klp10A overexpression may impair cellular organization due to the depolymerization of microtubules. Another common phenotype of Patronin RNAi and Klp10A overexpression is the wing blister. Wing blisters result from the failure of precise apposition of dorsal and ventral wing blades during the pupal stage [50]. Analysis of pupal wings would be necessary to determine whether the apical defects caused by Patronin loss or Klp10A overexpression could contribute to the blister formation.
In addition to its role in the maintenance of cell organization, Patronin is required for organ size. Our data show that the wing size defect by Patronin knockdown is mainly through cell death rather than defective cell proliferation. An interesting question is when cells die by Patronin loss during wing development. Ectopic Dcp-1 stain induced by Patronin RNAi or Klp10A overexpression suggests that Dcp-1 activation occurs in larval wing discs. Interestingly, although Diap1 overexpression can rescue the adult wing phenotype of Klp10A overexpression, it could not inhibit Dcp-1 activation in wing discs. In contrast, p35 overexpression not only suppresses the adult wing size defects of Klp10A overexpression but also reduces Dcp-1 activation in wing discs. It is possible that Diap1 may inhibit Dcp-1 activation in the pupal wing to recover the normal size of adult wings. In normal wing development, larval wing discs undergo dynamic morphogenetic changes including the wing blade apposition during the pupal stage, leading to adult wings. Patronin and Klp10A may play a role in morphogenesis and cell survival during this period. Uneven cell death in pupal wings may also contribute to imprecise apposition of wing blades, resulting in blister formation. It would be necessary to analyze pupal wings to find specific cellular defects and the time course for Dcp-1 activation in Patronin RNAi or Klp10A overexpression and Diap1-dependent suppression.
Patronin-Klp10A interaction and Hippo signaling
Our data suggest that the role of Patronin for microtubule stability might be related to the regulation of cell survival and organ size. Based on the observed relationships of Patronin with Klp10A and the Hippo pathway, we propose that Patronin plays dual roles in antagonizing the function of Klp10A and Hippo signaling. First, Patronin and Klp10A act antagonistically to maintain their protein levels. Since reduced Patronin levels by Klp10A overexpression cannot be suppressed by Diap1 or p35 overexpression (Fig. S13b′, d′, f′), the interaction between Patronin and Klp10A may be an upstream process of cell death regulation.
Second, Patronin may inhibit cell death by interacting with the Hippo pathway. We have shown that heterozygote mutations in hippo (hpo), warts (wts), and ex suppress Patronin RNAi phenotypes. Furthermore, Patronin RNAi wing phenotypes are restored by Yki overexpression, supporting a connection between Patronin and Hippo signaling. An intriguing question is how Patronin is related to Hippo signaling. One possibility is that Patronin might act as a modulator of the Hippo signaling by interacting with multiple Hippo pathway components. In particular, Patronin shows physical interactions with Mer and Ex. Furthermore, Patronin RNAi phenotypes in the wing are strongly suppressed by reducing Ex or Mer, suggesting a role of Patronin in an upstream of Hpo. The proposed Patronin function in inhibiting cell death through Hippo signaling is supported by the observation that Dcp-1 suppression by the wtsP2/+ heterozygote mutation.
Although Patronin RNAi phenotypes are suppressed by Yki overexpression, we could not clearly detect a change of Yki levels in Patronin-depleted wing discs. This is also consistent with the lack of change in the level of Diap1-lacZ in the wing disc. An intriguing question is how Patronin RNAi show little reduction of Yki activation in wing discs, despite strong evidence for genetic interactions between Patronin and Hippo pathway genes in the adult wing. It is possible that the reduction of activated Yki level may be too subtle for reliable detection in Patronin RNAi wing discs. Alternatively, because total Yki and p-Yki levels are reduced in adult Patronin heterozygote mutant tissues, changes in Yki by Patronin RNAi may occur in the pupal wing.
Recent studies have shown that CAMSAP proteins are involved in organizing ncMT in epithelial cells [15, 16]. Interestingly, Camsap2 or 3 mutant mice show smaller bodies [16, 51, 52], implying growth defects. It would be interesting to see whether potential growth defects in Camsap mutant mice are related to the control of cell survival and Hippo signaling during development.
Change history
12 August 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41418-020-00603-z
References
Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–24.
Akhmanova A, Hoogenraad CC. Microtubule minus-end-targeting proteins. Curr Biol. 2015;25:R162–71.
Wu J, Akhmanova A. Microtubule-organizing centers. Annu Rev Cell Dev Biol. 2017;33:51–75.
Martin M, Akhmanova A. Coming into focus: mechanisms of microtubule minus-end organization. Trends Cell Biol. 2018;28:574–88.
Akhmanova A, Steinmetz MO. Microtubule minus-end regulation at a glance. J Cell Sci. 2019;132:jcs227850.
Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–21.
Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–74.
Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, et al. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–70.
Wrighton KH. Cytoskeleton: keeping minus ends stable. Nat Rev Mol Cell Biol. 2010;11:816.
O’Rourke BP, Sharp DJ. Microtubule dynamics: patronin, protector of the minus end. Curr Biol. 2011;21:R77–9.
Wang H, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J Cell Biol. 2013;203:35–46.
Nashchekin D, Fernandes AR, St Johnston D. Patronin/Shot cortical foci assemble the noncentrosomal microtubule array that specifies the Drosophila anterior-posterior axis. Dev Cell. 2016;38:61–72.
Takeichi M, Toya M. Patronin takes a Shot at polarity. Dev Cell. 2016;38:12–3.
Khanal I, Elbediwy A, Diaz de la Loza Mdel C, Fletcher GC, Thompson BJ. Shot and patronin polarise microtubules to direct membrane traffic and biogenesis of microvilli in epithelia. J Cell Sci. 2016;129:2651–9.
Tanaka N, Meng W, Nagae S, Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci USA. 2012;109:20029–34.
Toya M, Kobayashi S, Kawasaki M, Shioi G, Kaneko M, Ishiuchi T, et al. CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc Natl Acad Sci USA. 2016;113:332–7.
Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34.
Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33.
Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–8.
Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–27.
Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol. 2010;20:R574–82.
Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–24.
Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22:695–702.
Gaspar P, Tapon N. Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol. 2014;31:74–83.
Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17.
Xu HM, Gutmann DH. Merlin differentially associates with the microtubule and actin cytoskeleton. J Neurosci Res. 1998;51:403–15.
Muranen T, Gronholm M, Lampin A, Lallemand D, Zhao F, Giovannini M, et al. The tumor suppressor merlin interacts with microtubules and modulates Schwann cell microtubule cytoskeleton. Hum Mol Genet. 2007;16:1742–51.
Smole Z, Thoma CR, Applegate KT, Duda M, Gutbrodt KL, Danuser G, et al. Tumor suppressor NF2/Merlin is a microtubule stabilizer. Cancer Res. 2014;74:353–62.
Bensenor LB, Barlan K, Rice SE, Fehon RG, Gelfand VI. Microtubule-mediated transport of the tumor-suppressor protein Merlin and its mutants. Proc Natl Acad Sci USA. 2010;107:7311–6.
Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–46.
Derivery E, Seum C, Daeden A, Loubery S, Holtzer L, Julicher F, et al. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature. 2015;528:280–5.
Laycock JE, Savoian MS, Glover DM. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J Cell Sci. 2006;119:2354–61.
McCartney BM, Fehon RG. Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophila homologues of moesin and the neurofibromatosis 2 tumor suppressor, merlin. J Cell Biol. 1996;133:843–52.
Bhat MA, Izaddoost S, Lu Y, Cho KO, Choi KW, Bellen HJ. Discs Lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell. 1999;96:833–45.
Ikmi A, Gaertner B, Seidel C, Srivastava M, Zeitlinger J, Gibson MC. Molecular evolution of the Yap/Yorkie proto-oncogene and elucidation of its core transcriptional program. Mol Biol Evol. 2014;31:1375–90.
Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–9.
Chung HL, Augustine GJ, Choi KW. Drosophila Schip1 links expanded and Tao-1 to regulate hippo signaling. Dev Cell. 2016;36:511–24.
Radford SJ, Harrison AM, McKim KS. Microtubule-depolymerizing kinesin KLP10A restricts the length of the acentrosomal meiotic spindle in Drosophila females. Genetics. 2012;192:431–40.
Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37.
Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–88.
Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–100.
Hong ST, Choi KW. TCTP directly regulates ATM activity to control genome stability and organ development in Drosophila melanogaster. Nat Commun. 2013;4:2986.
Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15.
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6.
Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125:2561–9.
Skouloudaki K, Christodoulou I, Khalili D, Tsarouhas V, Samakovlis C, Tomancak P, et al. Yorkie controls tube length and apical barrier integrity during airway development. J Cell Biol. 2019;218:2762–81.
McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–24.
Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27.
Fristrom DK, Rickoll WL. The morphogenesis of imaginal discs of Drosophila. Insect Ultrastructure. New York, NY, USA: Springer Science+ Business Media; 1982. p. 247–77.
Zusman S, Patel-King RS, Ffrench-Constant C, Hynes RO. Requirements for integrins during Drosophila development. Development. 1990;108:391–402.
Muroyama A, Terwilliger M, Dong B, Suh H, Lechler T. Genetically induced microtubule disruption in the mouse intestine impairs intracellular organization and transport. Mol Biol Cell. 2018;29:1533–41.
Pongrakhananon V, Saito H, Hiver S, Abe T, Shioi G, Meng W, et al. CAMSAP3 maintains neuronal polarity through regulation of microtubule stability. Proc Natl Acad Sci USA. 2018;115:9750–5.
Acknowledgements
We are grateful to all who kindly provided reagents and fly stocks described in “Materials and methods”, especially, N. Tapon, R. Fehon, G. Halder, D. St. Johnston, D. Glover, K. McKim. We also thank K.-O. Cho, M. Takeichi, C. Desplan, and our lab members for their valuable comments. This research was supported by NRF-2014K1A1A2042982 and NRF-2017R1A2B3007516 through the National Research Foundation of Korea, funded by the Korean Ministry of Education, Science & Technology.
Author information
Authors and Affiliations
Contributions
D-WY and K-WC designed the study, and D-WY performed experiments. D-WY and K-WC analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E. Baehrecke
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, DW., Choi, KW. Suppression of Patronin deficiency by altered Hippo signaling in Drosophila organ development. Cell Death Differ 28, 233–250 (2021). https://doi.org/10.1038/s41418-020-0597-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0597-x