Abstract
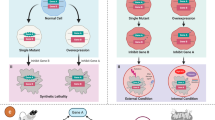
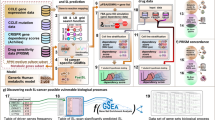
Since trastuzumab was approved in 2012 for the first-line treatment of gastric cancer (GC), no significant advancement in GC targeted therapies has occurred. Synthetic lethality refers to the concept that simultaneous dysfunction of a pair of genes results in a lethal effect on cells, while the loss of an individual gene does not cause this effect. Through exploiting synthetic lethality, novel targeted therapies can be developed for the individualized treatment of GC. In this study, we proposed a computational strategy named Gastric cancer Specific Synthetic Lethality inference (GSSL) to identify synthetic lethal interactions in GC. GSSL analysis was used to infer probable synthetic lethality in GC using four accessible clinical datasets. In addition, prediction results were confirmed by experiments. GSSL analysis identified a total of 34 candidate synthetic lethal pairs, which included 33 unique targets. Among the synthetic lethal gene pairs, TP53-CHEK1 was selected for further experimental validation. Both computational and experimental results indicated that inhibiting CHEK1 could be a potential therapeutic strategy for GC patients with TP53 mutation. Meanwhile, in vitro experimental validation of two novel synthetic lethal pairs TP53-AURKB and ARID1A-EP300 further proved the universality and reliability of GSSL. Collectively, GSSL has been shown to be a reliable and feasible method for comprehensive analysis of inferring synthetic lethal interactions of GC, which may offer novel insight into the precision medicine and individualized treatment of GC.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data and access to all databases used in this study are publicly available.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J Clinicians. 2018;68:394–424.
Zhang PH, Zheng ZG, Ling L, Yang XH, Zhang N, Wang X, et al. w09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS-RAF1-MAP2K-MAPK1/3 pathway. Autophagy. 2017;13:1093–112.
Chung HC, Bang YJ, Fuchs CS, Qin SK, Satoh T, Shitara K, et al. First-line pembrolizumab/placebo plus trastuzumab and chemotherapy in HER2-positive advanced gastric cancer: KEYNOTE-811. Future Oncol. 2021;17:491–501.
Wang DS, Liu ZX, Lu YX, Bao H, Wu X, Zeng ZL, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut. 2019;68:1152–61.
Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–8.
Fong PC, Boss DS, Yap TA, Tutt A, Wu PJ, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34.
Yeoh KG, Tan PT. Mapping the genomic diaspora of gastric cancer. Nat Rev Cancer. 2022;22:71–84.
Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102.
Zecchini V, Frezza C. Metabolic synthetic lethality in cancer therapy. Biochimica Et Biophysica Acta-Bioenerg. 2017;1858:723–31.
Muller FL, Aquilanti EA, DePinho RA. Collateral lethality: a new therapeutic strategy in oncology. Trends Cancer. 2015;1:161–73.
Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158:1199–209.
Wang J, Zhang Q, Han J, Zhao Y, Zhao C, Yan B, et al. Computational methods, databases and tools for synthetic lethality prediction. Brief Bioinform. 2022;23:bbac106.
Sinha S, Thomas D, Chan S, Gao Y, Brunen D, Torabi D, et al. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat Commun. 2017;8:13.
Lee JS, Das A, Jerby-Arnon L, Arafeh R, Auslander N, Davidson M, et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nat Commun. 2018;9:12.
Das S, Deng X, Camphausen K, Shankavaram U. DiscoverSL: an R package for multi-omic data driven prediction of synthetic lethality in cancers. Bioinformatics. 2019;35:701–2.
Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–U217.
Lei ZD, Tan IB, Das K, Deng NT, Zouridis H, Pattison S, et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–65.
Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, et al. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. Bmc Cancer. 2020;20:10.
Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8.
Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–5.
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16.
Bashashati A, Haffari G, Ding JR, Ha G, Lui K, Rosner J, et al. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol. 2012;13:14.
Wu GM, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:23.
He CY, Qiu MZ, Yang XH, Zhou DL, Ma JJ, Long YK, et al. Classification of gastric cancer by EBV status combined with molecular profiling predicts patient prognosis. Clin Transl Med. 2020;10:353–62.
Ge S, Xia X, Ding C, Zhen B, Zhou Q, Feng J, et al. A proteomic landscape of diffuse-type gastric cancer. Nat Commun. 2018;9:1012.
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82.
Lee YS, Cho YS, Lee GK, Lee S, Kim YW, Jho S, et al. Genomic profile analysis of diffuse-type gastric cancers. Genome Biol. 2014;15:R55.
Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Jeong SH, Park M, Park SY, Park J, Kim TH, Lee YJ, et al. Transcriptome analysis and the prognostic role of NUDC in diffuse and intestinal gastric cancer. Technol Cancer Res Treat. 2021;20:15330338211019501.
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83.
Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–2.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou SB, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–58.
Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–15.
Huang JK, Jia TQ, Carlin DE, Ideker T. pyNBS: a Python implementation for network-based stratification of tumor mutations. Bioinformatics. 2018;34:2859–61.
Tan IB, Ivanova T, Lim KH, Ong CW, Deng NT, Lee J, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–U551.
Cao J, Gong J, Li XH, Hu ZX, Xu YJ, Shi H, et al. Unsupervised hierarchical clustering identifies immune gene subtypes in gastric cancer. Front Pharmacol. 2021;12:12.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7.
Corsello SM, Bittker JA, Liu ZH, Gould J, McCarren P, Hirschman JE, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017;23:405–8.
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082.
Han YA, Yu GC, Sarioglu H, Caballero-Martinez A, Schlott F, Ueffing M, et al. Proteomic investigation of the interactome of FMNL1 in hematopoietic cells unveils a role in calcium-dependent membrane plasticity. J Proteom. 2013;78:72–82.
Yu, G. Gene Ontology semantic similarity analysis using GOSemSim. In: Kidder BL, editor. Stem cell transcriptional networks: methods and protocols, 2nd Edition. Totowa, USA: Humana Press Inc: 2020. pp. 207–15.
Ritchie ME, Phipson B, Wu D, Hu YF, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:13.
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1:417–25.
Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80.
Azuaje F. Computational models for predicting drug responses in cancer research. Brief Bioinforma. 2017;18:820–9.
Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9:3.
Altelaar AFM, Munoz J, Heck AJR. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48.
Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32.
Yamamoto M, Maehara Y, Oda S, Ichiyoshi Y, Kusumoto T, Sugimachi K. The p53 tumor suppressor gene in anticancer agent-induced apoptosis and chemosensitivity of human gastrointestinal cancer cell lines. Cancer Chemother Pharmacol. 1999;43:43–49.
Miller JJ, Orvain C, Jozi S, Clarke RM, Smith JR, Blanchet A, et al. Multifunctional compounds for activation of the p53-Y220C mutant in cancer. Chem-a Eur J. 2018;24:17734–42.
Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32:2866–8.
Rosales KR, Reid MA, Yang Y, Tran TQ, Wang WI, Lowman X, et al. TIPRL Inhibits protein phosphatase 4 activity and promotes H2AX phosphorylation in the DNA damage response. PLoS ONE. 2015;10:e0145938.
Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–72.
Pon JR, Marra MA. Driver and passenger mutations in cancer. In: Abbas AK, Galli SJ, Howley PM, editors. Annual review of pathology: mechanisms of disease. Vol. 10. Annual Reviews: Palo Alto; 2015. pp. 25–50.
Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat Rev Drug Discov. 2020;19:23–38.
O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–23.
Gemble S, Wardenaar R, Keuper K, Srivastava N, Nano M, Mace AS, et al. Genetic instability from a single S phase after whole-genome duplication. Nature. 2022;604:146–51.
Gali-Muhtasib H, Kuester D, Mawrin C, Bajbouj K, Diestel A, Ocker M, et al. Thymoquinone triggers inactivation of the stress response pathway sensor CHEK1 and contributes to apoptosis in colorectal cancer cells. Cancer Res. 2008;68:5609–18.
Brown C. An elusive cancer target. Nature. 2016;537:S106–8.
Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA. 2021;71:264–79.
Gadhikar MA, Sciuto MR, Alves MVO, Pickering CR, Osman AA, Neskey DM, et al. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Therapeutics. 2013;12:1860–73.
Alcaraz-Sanabria A, Nieto-Jimenez C, Corrales-Sanchez V, Serrano-Oviedo L, Andres-Pretel F, Montero JC, et al. Synthetic lethality interaction between aurora kinases and CHEK1 inhibitors in ovarian cancer. Mol Cancer Therapeutics. 2017;16:2552–62.
Ohashi S, Kikuchi O, Nakai Y, Ida T, Saito T, Kondo Y, et al. Synthetic lethality with trifluridine/tipiracil and checkpoint kinase 1 inhibitor for esophageal squamous cell carcinoma. Mol Cancer Therapeutics. 2020;19:1363–72.
Hubackova S, Davidova E, Boukalova S, Kovarova J, Bajzikova M, Coelho A, et al. Replication and ribosomal stress induced by targeting pyrimidine synthesis and cellular checkpoints suppress p53-deficient tumors. Cell Death Dis. 2020;11:16.
Origanti S, Cai SR, Munir AZ, White LS, Piwnica-Worms H. Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene. 2013;32:577–88.
Kogan-Sakin I, Tabach Y, Buganim Y, Molchadsky A, Solomon H, Madar S, et al. Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ. 2011;18:271–81.
Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–74.
Hong D, Infante J, Janku F, Jones S, Nguyen LM, Burris H, et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J Clin Oncol. 2016;34:1764–71.
Funding
This work was supported by The National Natural Science Foundation of China (81972206 and 82173215) and the Natural Science Foundation of Shanghai (22ZR1438800).
Author information
Authors and Affiliations
Contributions
Conceptualization, HG and RQ; methodology, HG, RQ, YZ, and XZ; validation, LZ, XT, and RQ; formal analysis, YX; investigation, CY, ZD, and TL; resources, SK and LZ; data curation, ZD and TL; writing—original draft preparation, HG, YZ, and LZ; writing—review and editing, WQ and CW; visualization, HG and RQ; supervision, ZZ and CZ; project administration, WQ and CW; funding acquisition, ZZ and CZ. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Geng, H., Qian, R., Zhong, Y. et al. Leveraging synthetic lethality to uncover potential therapeutic target in gastric cancer. Cancer Gene Ther 31, 334–348 (2024). https://doi.org/10.1038/s41417-023-00706-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-023-00706-y