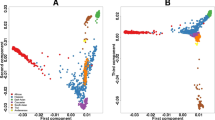
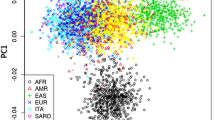
Abstract
Inter-individual variation of drug metabolising enzymes (DMEs) leads to variable efficacy of many drugs and even adverse drug responses. Consequently, it would be desirable to test variants of many DMEs before drug treatment. Inter-ethnic differences in frequency mean that the choice of SNPs to test may vary across population groups. Here we examine the utility of testing representative groups as a way of assessing what variants might be tested. We show that publicly available population information is potentially useful for determining loci for pre-treatment genetic testing, and for determining the most prevalent risk haplotypes in defined groups. However, we also show that the NHS England classifications have limitations for grouping for these purposes, in particular for people of African descent. We conclude: (1) genotyping of hospital patients and people from the hospital catchment area confers no advantage over using samples from appropriate existing ethnic group collections or publicly available data, (2) given the current NHS England Black African grouping, a decision as to whether to test, would have to apply to all patients of recent Black African ancestry to cover reported risk alleles and (3) the current scarcity of available genome and drug effect data from Africans is a problem for both testing and treatment decisions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Code availability
Data pertaining to the 1000G SNPs were extracted using VCFtools version 0.1.13 (https://vcftools.github.io/index.html), from the 1000G Phase 3 VCF files for the relevant chromosomes (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/). For systematic and automated haplotype analysis of large SNP data, we developed an R-based tool to convert PLINK PED/MAP files to PHASE input, and to summarise haplotype inference results in multiple groups from PHASE output. This tool is now publicly available on Github (https://github.com/nansari-pour/PLINKtoPHASE).
References
Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genom Hum Genet. 2006;7:223–45.
Daly AK. Pharmacogenetics: a general review on progress to date. Br Med Bull. 2017;124:65–79.
Browning LA, Kruse JA. Hemolysis and methemoglobinemia secondary to rasburicase administration. Ann Pharmacother. 2005;39:1932–5.
Khan S, Mandal RK, Elasbali AM, Dar SA, Jawed A, Wahid M, et al. Pharmacogenetic association between NAT2 gene polymorphisms and isoniazid induced hepatotoxicity: trial sequence meta-analysis as evidence. Biosci Rep. 2019;39:1–15.
Lonjou C, Borot N, Sekula P, Ledger N, Thomas L, Halevy S, et al. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genom. 2008;18:99–107.
Perry CM. Maraviroc: a review of its use in the management of CCR5-tropic HIV-1 infection. Drugs. 2010;70:1189–213.
Stehle S, Kirchheiner J, Lazar A, Fuhr U. Pharmacogenetics of oral anticoagulants: a basis for dose individualization. Clin Pharmacokinet. 2008;47:565–94.
Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015;25:33–41.
Davies EC, Green CF, Taylor S, Williamson PR, Mottram DR, Pirmohamed M. Adverse drug reactions in hospital in-patients: a prospective analysis of 3695 patient-episodes. PLoS ONE. 2009;4:e4439.
Dressler LG. Integrating personalized genomic medicine into routine clinical care: addressing the social and policy issues of pharmacogenomic testing. N C Med J. 2013;74:509–13.
Hovelson DH, Xue Z, Zawistowski M, Ehm MG, Harris EC, Stocker SL, et al. Characterization of ADME gene variation in 21 populations by exome sequencing. Pharmacogenet Genom. 2017;27:89.
Creemer OJ, Ansari-Pour N, Ekong R, Tarekegn A, Plaster C, Bains RK, et al. Contrasting exome constancy and regulatory region variation in the gene encoding CYP3A4: an examination of the extent and potential implications. Pharmacogenet Genom. 2016;26:255–70.
Gurwitz D, Motulsky AG. ‘Drug reactions, enzymes, and biochemical genetics’: 50 years later. Pharmacogenomics. 2007;8:1479–84.
Wilson JF, Weale ME, Smith AC, Gratrix F, Fletcher B, Thomas MG, et al. Population genetic structure of variable drug response. Nat Genet. 2001;29:265–9.
Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, et al. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–5.
Ferrell PB Jr, McLeod HL. Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics. 2008;9:1543–6.
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74.
Holmes MV, Shah T, Vickery C, Smeeth L, Hingorani AD, Casas JP. Fulfilling the promise of personalized medicine? Systematic review and field synopsis of pharmacogenetic studies. PLoS ONE. 2009;4:e7960.
Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37.
Francois AA, Nishida CR, de Montellano PRO, Phillips IR, Shephard EA. Human flavin-containing monooxygenase 2.1 catalyzes oxygenation of the antitubercular drugs thiacetazone and ethionamide. Drug Metab Dispos. 2009;37:178–86.
Veeramah KR, Thomas MG, Weale ME, Zeitlyn D, Tarekegn A, Bekele E, et al. The potentially deleterious functional variant flavin-containing monooxygenase 2*1 is at high frequency throughout sub-Saharan Africa. Pharmacogenet Genom. 2008;18:877–86.
Horsfall LJ, Zeitlyn D, Tarekegn A, Bekele E, Thomas MG, Bradman N, et al. Prevalence of clinically relevant UGT1A alleles and haplotypes in African populations. Ann Hum Genet. 2011;75:236–46.
Gammal RS, Court MH, Haidar CE, Iwuchukwu OF, Gaur AH, Alvarellos M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir prescribing. Clin Pharmacol Ther. 2016;99:363–9.
Bains RK. African variation at Cytochrome P450 genes: evolutionary aspects and the implications for the treatment of infectious diseases. Evol Med Public Health. 2013;2013:118–34.
Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genom Hum Genet. 2008;9:403–33.
Choudhury A, Aron S, Sengupta D, Hazelhurst S, Ramsay M. African genetic diversity provides novel insights into evolutionary history and local adaptations. Hum Mol Genet. 2018;27(R2):R209–R218.
Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–32.
de Man FM, Goey AKL, van Schaik RHN, Mathijssen RHJ, Bins S. Individualization of Irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 2018;57:1229–54.
Sugatani J, Mizushima K, Osabe M, Yamakawa K, Kakizaki S, Takagi H, et al. Transcriptional regulation of human UGT1A1 gene expression through distal and proximal promoter motifs: implication of defects in the UGT1A1 gene promoter. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:597–605.
Han FF, Guo CL, Yu D, Zhu J, Gong LL, Li GR, et al. Associations between UGT1A1*6 or UGT1A1*6/*28 polymorphisms and irinotecan-induced neutropenia in Asian cancer patients. Cancer Chemother Pharmacol. 2014;73:779–88.
Cui C, Shu C, Cao D, Yang Y, Liu J, Shi S, et al. UGT1A1*6, UGT1A7*3 and UGT1A9*1b polymorphisms are predictive markers for severe toxicity in patients with metastatic gastrointestinal cancer treated with irinotecan-based regimens. Oncol Lett. 2016;12:4231–7.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Ansari Pour N, Plaster CA, Bradman N. Evidence from Y-chromosome analysis for a late exclusively eastern expansion of the Bantu-speaking people. Eur J Hum Genet. 2013;21:423–9.
R-Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014.
Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9.
Lee CR, Pieper JA, Frye RF, Hinderliter AL, Blaisdell JA, Goldstein JA. Differences in flurbiprofen pharmacokinetics between CYP2C9*1/*1, *1/*2, and *1/*3 genotypes. Eur J Clin Pharmacol. 2003;58:791–4.
Perini JA, Vianna-Jorge R, Brogliato AR, Suarez-Kurtz G. Influence of CYP2C9 genotypes on the pharmacokinetics and pharmacodynamics of piroxicam. Clin Pharmacol Ther. 2005;78:362–9.
Rettie AE, Haining RL, Bajpai M, Levy RH. A common genetic basis for idiosyncratic toxicity of warfarin and phenytoin. Epilepsy Res. 1999;35:253–5.
Tang C, Shou M, Mei Q, Rushmore TH, Rodrigues AD. Major role of human liver microsomal cytochrome P450 2C9 (CYP2C9) in the oxidative metabolism of celecoxib, a novel cyclooxygenase-II inhibitor. J Pharmacol Exp Ther. 2000;293:453–9.
Furuta T, Ohashi K, Kosuge K, Zhao XJ, Takashima M, Kimura M, et al. CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin Pharmacol Ther. 1999;65:552–61.
Goldstein JA, Faletto MB, Romkes-Sparks M, Sullivan T, Kitareewan S, Raucy JL, et al. Evidence that CYP2C19 is the major (S)-mephenytoin 4’-hydroxylase in humans. Biochemistry. 1994;33:1743–52.
Hirani VN, Raucy JL, Lasker JM. Conversion of the HIV protease inhibitor nelfinavir to a bioactive metabolite by human liver CYP2C19. Drug Metab Dispos. 2004;32:1462–7.
Inomata S, Nagashima A, Itagaki F, Homma M, Nishimura M, Osaka Y, et al. CYP2C19 genotype affects diazepam pharmacokinetics and emergence from general anesthesia. Clin Pharmacol Ther. 2005;78:647–55.
Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–23.
Tseng E, Walsky RL, Luzietti RA Jr, Harris JJ, Kosa RE, Goosen TC, et al. Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos. 2014;42:1163–73.
Phillips IR, Shephard EA. Drug metabolism by flavin-containing monooxygenases of human and mouse. Expert Opin Drug Metab Toxicol. 2017;13:167–81.
McDonagh EM, Boukouvala S, Aklillu E, Hein DW, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genom. 2014;24:409–25.
Innocenti F, Ratain MJ. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping. Pharmacogenomics. 2006;7:1211–21.
Acknowledgements
We thank all the sample donors who participated in this study and the UCLH clinicians, Aroon Hingorani, Alastair Forbes, Simon Woldman, Steve Hurel, Clare Dollery and others who helped us with access to patient volunteers in their clinics. We also thank Mark Thomas for access to some of the samples and Pieta Nosanea, Esther Williams, Sarah Steward and Ayele Tarekegn for help with sample collections from African and Indian volunteers in their native countries; Ranji Areseratnam for technical assistance. This research was funded by the University College London Hospitals Comprehensive Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
During this study NB had a controlling interest in a company interested in developing diagnostic technology to identify variation in drug metabolising enzymes to improve healthcare. Neither NB nor the company now have that objective. None of the other authors have any potential conflicts of interest to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ingram, C.J.E., Ekong, R., Ansari-Pour, N. et al. Group-based pharmacogenetic prediction: is it feasible and do current NHS England ethnic classifications provide appropriate data?. Pharmacogenomics J 21, 47–59 (2021). https://doi.org/10.1038/s41397-020-0175-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-020-0175-0