Abstract
Background
Multicystic dysplastic kidney (MCDK) is a common form of congenital kidney anomaly. The cause of MCDK is unknown. We investigated whether MCDK in children is linked to cytogenomic aberrations.
Methods
We conducted array comparative genomic hybridization (aCGH) in ten unrelated children with MCDK. The pattern of inheritance was determined by real-time PCR in patients and their biological parents.
Results
Pathogenic aberrations were detected in three patients: a deletion at 7p14.3 with a size of 2.07 Mb housing 12 genes, including BBS9 (Bardet–Biedl syndrome 9) and BMPER (BMP binding endothelial regulator); a duplication at 16p13.11p12.3 with a size of 3.28 Mb that included >20 genes; and monosomy X for a female patient. The deletion at 7p14.3 was inherited from the patient’s father, while the duplication at 16p13.11p12.3 was derived from the patient’s mother.
Conclusions
Up to 30% of patients with MCDK possess cytogenomic aberrations. BBS9 and BMPER variants have been reported to result in cystic kidney dysplasia, suggesting a possible pathogenic function for the deletion at 7p14.3 in children with MCDK. The duplication at 16p13.11p12.3 was not reported previously to associate with MCDK. Both variations were inherited from parents, indicating hereditary contributions in MCDK. Thus, aCGH is an informative tool to unravel the pathogenic mechanisms of MCDK.
Impact
-
Cytogenomic aberrations are common in children with MCDK.
-
Cytogenomic aberrations are inherited from parents, indicating hereditary contributions in MCDK.
-
aCGH is a valuable tool to reveal pathogenic mechanisms of MCDK.
Similar content being viewed by others
Introduction
Multicystic dysplastic kidney (MCDK) is a form of congenital anomaly of the kidney and urinary tract (CAKUT). CAKUT is a major cause of kidney failure in children, accounting for 30–50% of cases of end-stage kidney disease.1 Multiple lines of evidence, including the discovery of causative genes and intrafamilial CAKUT segregation, support the contribution of genetic factors to CAKUT in humans. A recent large-scale study by Verbitsky et al.2 reported the presence of a large size (≥100 kb) copy-number variations (CNVs) in 4.1% of patients with diverse forms of CAKUT recruited in the United States, Europe, and Brazil, demonstrating that genomic disorders represent a common etiology of CAKUT. However, the gene variants causing a discrete form of CAKUT such as MCDK remain incompletely defined. MCDK consists of macroscopic cysts with absent working kidney tissue and arises in 1 in 1000 to 1 in 4300 live births.3,4 MCDK is unilateral in most cases, but may affect both kidneys. Bilateral MCDK causes Potter syndrome (hypoplastic lungs, deformed limbs, widely separated eyes, broad nasal bridge, and low set ears). Histological examination of MCDK tissue demonstrates the presence of connective tissue and immature epithelium.3,4 MCDK is believed to result from either abnormal inductive interactions between the ureteric bud and the metanephric mesenchyme during embryonic kidney development or from intrauterine fetal urinary tract obstruction.5,6 Variants in genes critical for normal kidney development (the so-called renal developmental genes), including HNF1B, ACE, PAX2, REN, ROBO2, AGTR1, SALL1, and AGT genes, have been linked to pediatric MCDK.7,8,9,10 These observations indicate that genetic alterations may play a causative role in MCDK.
In addition to single variants of discrete genes, large cytogenetic defects, including the number of copies of a particular gene, insertions, deletions, and duplications of large DNA segments, are associated with CAKUT.11 CNVs are defined as any gain or loss of germline DNA. The characterization of DNA CNVs by array comparative genomic hybridization (aCGH) analysis has demonstrated to be highly valuable in revealing pathogenic mechanisms of CAKUT2,11. However, the pathogenic roles of cytogenomic aberrations in children with MCDK were not investigated well. Here, we report the results of a study using aCGH in ten pediatric patients with isolated MCDK.
Materials and methods
Patients
The study was approved by the Tulane University School of Medicine IRB#:150438-4. Informed consent was obtained from the parents of children and, where appropriate, assent was acquired from children themselves. Ten unrelated patients with MCDK (mean age 8.5 ± 1.1 years) were enrolled in the study after clinical diagnosis of MCDK was established by renal ultrasonography (US). Blood was obtained from MCDK patients and 20 pooled age-, race- and sex-matched controls (six black and five white males, five black and four white females). Buccal cells were obtained from patients’ biological parents. Children in the control group had US performed for kidney diseases different from CAKUT (e.g., minimal proteinuria and microscopic hematuria). Six MCDK patients were females and four males. Kidney function was estimated from plasma creatinine with Schwartz equation (height (cm) × 0.413/plasma creatinine (mg/dL)).12
DNA isolation
DNA was obtained from blood or from patients’ biological parents’ buccal cells as previously described.10
aCGH analysis
DNA was isolated and labeled using Agilent-recommended protocol. CGH was performed on an Agilent Microarray platform with 105k probes (Agilent Inc., Santa Clara, CA). Array image was acquired with an Agilent Array Scanner. Microarray data were analyzed using the Cytogenomics software package from Agilent Inc. Interpretation of detected CNVs was conducted according to the ACMG standards and guidelines revision 2013,13 and technical standards recommendation by ACMG and ClinGen.14 Information for clinical significance on reported cases was extracted from Databases of Genomic Variants (ClinVar) at www.ncbi.nim.nih.gov/clinvar and DECIPHER at www.decipher.sanger.ac.uk. Association analysis of gene function and clinical features is based on the information from Online Mendelian Inheritance in Man (OMIM at www.omim.org).
Real-time quantitative polymerase chase reaction (qPCR)
Primers for qPCR were designed to detect the relative copy number of targeted genes within the aberrations and to avoid any potential common genomic variants encountered in the general population reported in the Database of Genomic Variants. The SYBR green assay (Thermo Fisher Scientific) was performed on BMPER (BMP binding endothelial regulator) in the deleted region and XYLT1 (xylosyltransferase 1) in the duplicated region, using RPPH1 (ribonuclease P RNA component H1) and TERT (telomerase reverse transcriptase) genes as references. The relative copy number was calculated using the ∆∆Ct method compared to an unaffected human DNA sample. Primers used for qPCR were as follows: BMPER-forward: ctgtggtttgcaagaggaag and BMPER-reverse: atgtcttctgggggcactc; XYLT1-forward: caacgagtccagccatcc and XYLT1-reverse: cagagcttccagagcctaaac; TERT-E3-forward: tcccacgacgtagtccat and TERT-E3- reverse: cagaggtca-ggcagcatc; RNaseP-forward: ggagagtagtctgaattgggttatg and RNaseP-reverse: ggagcttggaaca-gactcac.
Results
Clinical findings
The mean age of MCDK patients was 8.5 ± 1.1 years and of children in the control group was 9.7 ± 0.9 years (Tables 1 and 2). Four MCDK patients were males and six females. Eight children were African American and two Caucasian. All children manifested normal blood pressure and renal function. In all children, family history was negative for MCDK or other anomalies of the kidney and urinary tract. MCDK was isolated in nine of ten cases and was associated with Turner’s syndrome in one of the patient. MCDK was unilateral in all children with a ratio of right vs. left MCDK 1:1 (Table 1). The contralateral kidney underwent proper compensatory hypertrophy in all instances. Mild hydronephrosis was identified in the contralateral kidney of one patient. US did not reveal any renal malformations in children from the control group. All biological parents had apparently normal phenotype and reported the absence of known kidney disease or abnormalities of the urinary system.
aCGH results
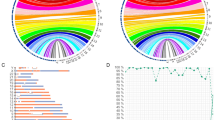
Three diverse pathogenic aberrations (a deletion, a duplication, and a numerical abnormality) were detected in three of ten patients (in two of nine patients with isolated MCDK and in a single patient with known Turner syndrome). The results are summarized in Table 3. No pathogenic aberrations were detected from the other seven patients using the laboratory standard cutoff at the resolution of 300 kb. The deletion detected from subject 3 was located at 7p14.3 with a size of 2.07 Mb. This alteration results in deletion of seven protein-coding genes, including BBS9, BMPER, and RP9 (retinitis pigmentosa 9) (Figs. 1 and 2a), as well as a pseudogene, RP9P (retinitis pigmentosa 9 pseudogene), and three copies of non-coding gene of NPSR (neuropeptide S receptor)-AS1 (antisense RNA 1). The duplication detected from subject 41 was located at 16p13.11p12.3 with a size of 3.28 Mb. There are >30 genes in this duplicated region, including 16 protein-coding genes, 18 microRNA (miRNA) genes in three clusters, and two pseudogenes. (Figs. 1 and 2b). The third aberration was monosomy X (Fig. 1) from a female patient (subject 22). The data of the laboratory analysis are shown in Table 3.
a Alteration regions and their genetic contents. Deletion at 7p14.3 from subject 3. Red arrows: genes in the detected regions with function possibly contributing to clinical phenotype (please see “Discussion” for details) or disease causing. b Duplication at 16p13.11p12.3 from subject 41. Red arrows: genes in the detected regions with function possibly contributing to clinical phenotype (please see discussion for details) or disease causing. Rectangles: clusters of microRNA genes that may play roles in the regulation of gene expression.
qPCR results
The results from qPCR assay confirmed the findings from aCGH of the deletion and duplication. qPCR studies of the two families showed that the deletion at 7p14.3 was inherited from the patient’s father (Fig. 3a), while the duplication at 16p13.11p12.3 was derived from the patient’s mother (Fig. 3b). The monosomy X is a known product of meiotic nondisjunction and no further confirmation study was carried.
Discussion
The current study identified novel cytogenomic aberrations in children with isolated MCDK. The overall prevalence of cytogenomic aberrations was ~33% (three of ten patients). The prevalence of cytogenomic aberrations in patients with isolated MCDK in this study was ~22% (two of nine). These findings are in line with other reports showing that molecular diagnosis due to a copy-number disorder can be established in 4.1–14.5% of children with diverse forms of CAKUT.2,11 Relative enrichment for CNVs in our analysis (22%) could be due to the study of a single discrete form of CAKUT such as MCDK rather than diverse forms of CAKUT. Our present findings support the role of genetic factors in the pathogenesis of MCDK. Understanding the genetic architecture of MCDK as a discrete form of CAKUT has important implications for the development of preventive and therapeutic interventions that aim to mitigate the associated cardiovascular comorbidities and curtail the progression of kidney disease. In addition, the identification of CNVs helps to identify novel intracellular pathways that are implicated in the pathogenesis of MCDK and to provide molecular diagnosis, thus establishing the etiology of MCDK.
The deletion at 7p14.3 results in the deletion of BBS9 and BMPER genes. Mutations in BBS9 cause Bardet–Biedl syndrome (BBS, OMIM 209900), a rare autosomal-recessive ciliopathy distinguished by mental retardation, polydactyly, obesity, retinitis pigmentosa, and CAKUT.15,16 Renal manifestations of BBS include collecting duct microcysts rather than macrocysts observed in MCDK.15,16 In the Databases of Decipher and ClinVar, ten patients with 7p14.3 deletion were reported with deletions in size from 200 to 2100 kb, and all these deletions have BBS9 and BMPER involved. Six of these patients showed clinical features of intellectual disability, autistic behavior, and developmental delay. One patient also manifested unilateral renal hypoplasia. Our patient with the 7p14.3 deletion (subject 3 in Tables 1 and 3) has clinical features of MCDK and autistic features, similar to this case. It is worth noting that the clinical features of these patients are different from that of BBS patients, suggesting that the phenotype of 7p14.3 deletion may be due to more complex mechanism of multiple gene deletions rather than to loss of function of a single BBS9 gene.
Animal studies demonstrate that mice deficient in a related gene, BBS4 (Bbs4−/−), exhibit renal glomerular macrocysts.17 These findings underscore the importance of BBS spectrum genes in cystogenesis in mammals. Mutations in BMPER, which encodes the bone morphogenetic protein (BMP)-binding endothelial cell precursor-derived regulator, cause diaphanospondylodysostosis (DSD, OMIM 608022), a rare autosomal-recessive disease characterized by aberrant vertebral segmentation and a small chest. Renal findings in DSD include nephroblastomatosis with cystic kidneys.18,19 Bmp4 is a member of the transforming growth factor-β family and is essential for normal kidney organogenesis. It inhibits ectopic outgrowth of the ureteric bud and promotes elongation of ureteric bud-derived ureter in mice.20 In addition, recombinant BMP4 induces cell apoptosis during early stages of kidney formation.21 Inhibition of the BMP signaling is critical for survival and proliferation of the nephron progenitor cells in the metanephric mesenchyme. Of interest, kidneys of newborn Bmp4+/− mice contain multicystic dysplastic areas.22 In humans, BMP4 variants are associated with renal hypodysplasia, defined as reduced renal size and/or abnormal formation of the kidney tissue during renal organogenesis.23 Collectively, disruption of BMPER/BMP signaling is linked to variable types of cystogenesis in both mice and humans.
The duplication at 16p13.11p12.3 spans over the region for 16p13.11 recurrent microduplication locus and extends to its downstream region. More than 20 genes are located within the duplicated region, including ten miRNA genes, and disease-associated genes such as NDE1, MYH11, ABCC6, and XYLT1. Two pseudogenes, PKD1P1 and ABCC6P1, are located within the duplication. PKD1P1 is a pseudogene for PKD1 (polycystic kidney disease 1). PKD1 mutations result in an autosomal-dominant polycystic kidney disease in humans. The PKD1P1 shares a 97.7% sequence identity with the genuine PKD1. PKD1P1 is expressed during the early stages of embryogenesis.24 However, its function has not been determined yet. Recent reports indicate that pseudogenes might affect the expression of their parental genes by diverting miRNAs away from corresponding parental mRNA.25 If this is the case for PDK1P1, an extra copy of PDK1P1 might have impacts on the expression of PKD1. ABCC6P1 is a pseudogene for ABCC6 (ATP binding cassette subfamily C member 6). Mutation of ABCC6 results in pseudoxanthoma elasticum, an inherited disease characterized by calcification of arteries and kidney tissue. Both genes are located in the duplicated region. ABCC6 is expressed in the kidney, suggesting its possible role in kidney development or function.24,26 The duplication will add extra one copy of ABCC6P1 and possibly alter the expression level of this gene in the fetal kidney. In addition, there are ~18 copies of miRNA genes within the duplicated region. Increasing copy number of miRNA genes may have an impact on the regulation of gene expression as well. Therefore, the pathogenic role of ABCC6P1 duplication in MCDK cannot be excluded. Both the deletion at 7p14.3 and the duplication at 16p13.11p12.3 were inherited from each patient’s phenotypically normal corresponding parents. These two cytogenomic aberrations have not been reported previously as CNVs from the general population, suggesting that they may be specific to MCDK.
Identification of CNVs in the minority of children with MCDK in this study is consistent with such general characteristics of CAKUT as incomplete penetrance and variable expressivity of disease. Additional mechanism explaining the lack of identifiable CNVs in children with MCDK include epigenetic imprinting and unmarked single-nucleotide variants in the aberration regions. Detailed analysis of genes located within identified aberrations will allow to better elucidate genetic mechanisms of MCDK. In this regard, next-generation sequencing, including whole-exome sequencing, should improve the discovery of novel causative genes in patients with MCDK and their families.
In summary, the current study describes two novel candidates of cytogenomic alterations as MCDK susceptibility loci. Unilateral renal defect was shown in two of seven patients with 7p14.3 deletion (including one case described in this study), suggesting that it is a common clinical feature of 7p14.3 deletion. We report the novel finding of MCDK association with the 16p13.11 duplication. We explored the mechanisms for several genes located within the identified cytogenomic alterations as possible genetic drivers for MCDK in children. Therefore, these results provide significant insight into the genomic landscape of MCDK in humans.
References
Song, R. & Yosypiv, IV. Genetics of congenital anomalies of the kidney and urinary tract. Pediatr. Nephrol. 26, 353–364 (2011).
Verbitsky, M. et al. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat. Genet. 51, 117–127 (2019).
Schreuder, M. F., van Wijk, J. A. & Westland, R. Unilateral multicystic dysplastic kidney: a meta-analysis of observational studies on the incidence, associated urinary tract malformations and the contralateral kidney. Nephrol. Dial. Transplant. 24, 1810–1818 (2009).
Sarhan, O. et al. Multicystic dysplastic kidney: impact of imaging modality selection on the initial management and prognosis. J. Pediatr. Urol. 10, 645–649 (2014).
Cussen, L. J. & Felson, B. The hydronephrotic type of unilateral congenital multicystic disease of the kidney. Semin. Roentgenol. 10, 113–123 (1975).
Kitagawa, H. et al. Early bladder wall changes after creation of obstructive uropathy in fetal lamb. Pediatr. Surg. Int. 22, 875–879 (2006).
Brugnara, M., Fanos, V., Franchini, M. & Zaffanello, M. TCF2 gene mutation leads to nephro-urological defects of unequal severity: an open question. Med. Sci. Monit. 14, RA78–RA86 (2008).
Fletcher, J. et al. Multicystic dysplastic kidney and variable phenotype in a family with a novel deletion mutation of PAX2. J. Am. Soc. Nephrol. 16, 2754–2761 (2005).
Hwang, D. Y. et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 85, 1429–1433 (2014).
Song, R. & Yosypiv I. V. Sequence variants in the renin-angiotensin system genes are associated with isolated multicystic dysplastic kidney in children. Pediatr. Res. https://doi.org/10.1038/s41390-020-01255-y (2020).
Sanna-Cherchi, S. et al. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet. 91, 987–997 (2012).
Schwartz, G. J. & Work, D. F. Measurement and estimation of GFR in children and adolescents. J. Am. Soc. Nephrol. 4, 1832–1843 (2009).
South, S. T. et al. For the Working Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet. Med. 15, 901–909 (2013).
Riggs, E. R. et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 22, 245–257 (2020).
Imhoff, O. et al. Bardet-Biedl syndrome: a study of the renal and cardiovascular phenotypes in a French Cohort. Clin. J. Am. Soc. Nephrol. 6, 22–29 (2011).
Nishimura, D. Y. et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am. J. Hum. Genet. 77, 1021–1033 (2005).
Guo, D. F. et al. Inactivation of Bardet-Biedl syndrome genes causes kidney defects. Am. J. Physiol. Ren. Physiol. 300, F574–F580 (2011).
Prefumo, F. et al. A newly recognized autosomal recessive syndrome with abnormal vertebral ossification, rib abnormalities, and nephrogenic rests. Am. J. Med. Genet. 120A, 386–388 (2003).
Zong, Z. et al. BMPER variants associated with a novel, attenuated subtype diaphanospondylodysostosis. J. Hum. Genet. 60, 743–747 (2015).
Fogo, A., Hogan, B. L., Ichikawa, I., Miyazaki, Y. & Oshima, K. Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J. Clin. Investig. 105, 863–873 (2000).
Michos, O. et al. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signaling during kidney branching morphogenesis. Development 134, 2397–2405 (2007).
Miyazaki, Y., Oshima, K., Fogo, A. & Ichikawa, I. Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int. 63, 835–844 (2003).
Weber, S. et al. SIX2 and BMP4 mutations associate with anomalous kidney development. J. Am. Soc. Nephrol. 19, 891–903 (2008).
Szabo, L. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16, 1–26 (2015).
An, Y., Furber, K. L. & Ji, S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell. Mol. Med. 21, 185–192 (2017).
Duff, M. O. et al. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 521, 376–379 (2015).
Acknowledgements
This work was supported by NIH Grant DK-071699 to I.V.Y.
Author information
Authors and Affiliations
Contributions
Each author has met the Pediatric Research authorship requirements. T.-J.C., I.V.Y., A.J. and R.S. had substantial contributions to conception and design, acquisition of data, analysis, and interpretation of data. T.-J.C. drafted the article. I.V.Y., T.-J.C. and R.S. revised thhe article for important intellectual content. T.J.C., R.S., A.J. and I.V.Y. approved the submitted version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Patient consent
Informed consent was obtained from the parents of children and, when appropriate, assent was acquired from children themselves.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, TJ., Song, R., Janssen, A. et al. Cytogenomic aberrations in isolated multicystic dysplastic kidney in children. Pediatr Res 91, 659–664 (2022). https://doi.org/10.1038/s41390-021-01476-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01476-9