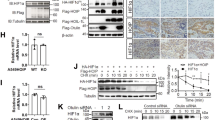
Abstract
Mutations in E3 ubiquitin ligase UBE3B have been linked to Kaufman Oculocerebrofacial Syndrome (KOS). Accumulating evidence indicates that UBE3B may play an important role in cancer. However, the precise role of UBE3B in cancer and the underlying mechanism remain largely uncharted. Here, we reported that UBE3B is an E3 ligase for hypoxia-inducible factor 2α (HIF-2α). Mechanically, UBE3B physically interacts with HIF-2α and promotes its lysine 63 (K63)-linked polyubiquitination, thereby inhibiting the Von Hippel-Lindau (VHL) E3 ligase complex-mediated HIF-2α degradation. UBE3B depletion inhibits breast cancer cell proliferation, colony formation, migration, and invasion in vitro and suppresses breast tumor growth and lung metastasis in vivo. We further identified K394, K497, and K503 of HIF-2α as key ubiquitination sites for UBE3B. K394/497/503R mutation of HIF-2α dramatically abolishes UBE3B-mediated breast cancer growth and lung metastasis. Intriguingly, the protein levels of UBE3B are upregulated and positively correlated with HIF-2α protein levels in breast cancer tissues. These findings uncover a critical mechanism underlying the role of UBE3B in HIF-2α regulation and breast cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
UBE3B expression data in human breast tumors in the TCGA dataset were downloaded from the UCSC Cancer Browser (http://genome-cancer.ucsc.edu). UBE3B mRNA expression was queried in adjacent normal breast tissues and primary and metastatic breast tumors with different stages or grades. Kaplan–Meier survival analysis was performed in breast cancer patients who were divided by medium expression levels of UBE3B mRNA. Kaplan–Meier plotter (http://kmplot.com) was utilized for overall survival analysis for breast cancer patients who were divided according to UBE3B protein levels. All data generated or analyzed during this study are included in this article and its supplementary information files. Any additional data presented in this paper are available from the corresponding author upon request.
References
Kariminejad A, Ajeawung NF, Bozorgmehr B, Dionne-Laporte A, Molidperee S, Najafi K, et al. Kaufman oculo-cerebro-facial syndrome in a child with small and absent terminal phalanges and absent nails. J Hum Genet. 2017;62:465–71.
Flex E, Ciolfi A, Caputo V, Fodale V, Leoni C, Melis D, et al. Loss of function of the E3 ubiquitin-protein ligase UBE3B causes Kaufman oculocerebrofacial syndrome. J Med Genet. 2013;50:493–9.
Basel-Vanagaite L, Yilmaz R, Tang S, Reuter MS, Rahner N, Grange DK, et al. Expanding the clinical and mutational spectrum of Kaufman oculocerebrofacial syndrome with biallelic UBE3B mutations. Hum Genet. 2014;133:939–49.
Cheon S, Kaur K, Nijem N, Tuncay IO, Kumar P, Dean M, et al. The ubiquitin ligase UBE3B, disrupted in intellectual disability and absent speech, regulates metabolic pathways by targeting BCKDK. Proc Natl Acad Sci USA. 2019;116:3662–7.
Ambrozkiewicz MC, Borisova E, Schwark M, Ripamonti S, Schaub T, Smorodchenko A, et al. The murine ortholog of Kaufman oculocerebrofacial syndrome protein Ube3b regulates synapse number by ubiquitinating Ppp3cc. Mol Psychiatry. 2021;26:1980–95.
Braganza A, Li J, Zeng X, Yates NA, Dey NB, Andrews J, et al. UBE3B Is a Calmodulin-regulated, Mitochondrion-associated E3 Ubiquitin Ligase. J Biol Chem. 2017;292:2470–84.
Svilar D, Dyavaiah M, Brown AR, Tang JB, Li J, McDonald PR, et al. Alkylation sensitivity screens reveal a conserved cross-species functionome. Mol Cancer Res. 2012;10:1580–96.
Li K, Wang F, Yang ZN, Zhang TT, Yuan YF, Zhao CX, et al. TRIB3 promotes MYC-associated lymphoma development through suppression of UBE3B-mediated MYC degradation. Nat Commun. 2020;11:6316.
Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–32.
Chen Y, Liu M, Niu Y, Wang Y. Romance of the three kingdoms in hypoxia: HIFs, epigenetic regulators, and chromatin reprogramming. Cancer Lett. 2020;495:211–23.
Wang Y, Li G, Deng M, Liu X, Huang W, Zhang Y, et al. The multifaceted functions of RNA helicases in the adaptive cellular response to hypoxia: from mechanisms to therapeutics. Pharmacol Ther. 2021;221:107783.
Wang Y, Liu X, Huang W, Liang J, Chen Y. The intricate interplay between HIFs, ROS, and the ubiquitin system in the tumor hypoxic microenvironment. Pharmacol Ther. 2022;240:108303.
Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, et al. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1alpha. Blood. 1998;92:2260–8.
Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13.
Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–23.
Briggs KJ, Koivunen P, Cao S, Backus KM, Olenchock BA, Patel H, et al. Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell. 2016;166:126–39.
Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, et al. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–21.
Helczynska K, Larsson AM, Holmquist Mengelbier L, Bridges E, Fredlund E, Borgquist S, et al. Hypoxia-inducible factor-2alpha correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Res. 2008;68:9212–20.
Kaelin WG Jr. The VHL tumor suppressor gene: insights into oxygen sensing and cancer. Trans Am Clin Climatol Assoc. 2017;128:298–307.
Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–7.
Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–11.
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64.
Yu W, Hu C, Gao H. Advances of nanomedicines in breast cancer metastasis treatment targeting different metastatic stages. Adv Drug Deliv Rev. 2021;178:113909.
Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–66.
Gao WW, Xiao RQ, Zhang WJ, Hu YR, Peng BL, Li WJ, et al. JMJD6 licenses ERalpha-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 Co-activator complex. Mol Cell. 2018;70:340–57. e348
Shen HF, Zhang WJ, Huang Y, He YH, Hu GS, Wang L, et al. The dual function of KDM5C in both gene transcriptional activation and repression promotes breast cancer cell growth and tumorigenesis. Adv Sci. 2021;8:2004635.
Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602.
Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci. 2012;37:364–72.
Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–25.
Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–74.
Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker-Jensen DB, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 2018;25:631–40.
Vogl AM, Phu L, Becerra R, Giusti SA, Verschueren E, Hinkle TB, et al. Global site-specific neddylation profiling reveals that NEDDylated cofilin regulates actin dynamics. Nat Struct Mol Biol. 2020;27:210–20.
Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–52.
Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–206.
Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–5.
Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8.
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72.
Niu Y, Bao L, Chen Y, Wang C, Luo M, Zhang B, et al. HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res. 2020;80:964–75.
Chen Y, Zhang B, Bao L, Jin L, Yang M, Peng Y, et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Investig. 2018;128:1937–55.
Schodel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–5. S421-422
Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Investig. 2009;119:2160–70.
Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–70.
Ősz Á, Lánczky A, Győrffy B. Survival analysis in breast cancer using proteomic data from four independent datasets. Sci Rep. 2021;11:16787.
Bernassola F, Chillemi G, Melino G. HECT-type E3 ubiquitin ligases in cancer. Trends Biochem Sci. 2019;44:1057–75.
So CL, Saunus JM, Roberts-Thomson SJ, Monteith GR. Calcium signalling and breast cancer. Semin Cell Dev Biol. 2019;94:74–83.
Tarade D, Robinson CM, Lee JE, Ohh M. HIF-2alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2alpha-driven disease. Nat Commun. 2018;9:3359.
Paltoglou S, Roberts BJ. HIF-1alpha and EPAS ubiquitination mediated by the VHL tumour suppressor involves flexibility in the ubiquitination mechanism, similar to other RING E3 ligases. Oncogene. 2007;26:604–9.
Miyauchi Y, Kato M, Tokunaga F, Iwai K. The COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-alpha ubiquitination. J Biol Chem. 2008;283:16622–31.
Wallace EM, Rizzi JP, Han G, Wehn PM, Cao Z, Du X, et al. A small-molecule antagonist of HIF2alpha is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 2016;76:5491–500.
Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22.
Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat Genet. 2003;35:331–40.
Kim WY, Safran M, Buckley MR, Ebert BL, Glickman J, Bosenberg M, et al. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. EMBO J. 2006;25:4650–62.
Rattner A, Williams J, Nathans J. Roles of HIFs and VEGF in angiogenesis in the retina and brain. J Clin Invest. 2019;129:3807–20.
Kleszka K, Leu T, Quinting T, Jastrow H, Pechlivanis S, Fandrey J, et al. Hypoxia-inducible factor-2alpha is crucial for proper brain development. Sci Rep. 2020;10:19146.
Leu T, Fandrey J, Schreiber T. (H)IF applicable: promotion of neurogenesis by induced HIF-2 signalling after ischaemia. Pflugers Arch. 2021;473:1287–99.
Leiton CV, Chen E, Cutrone A, Conn K, Mellanson K, Malik DM, et al. Astrocyte HIF-2alpha supports learning in a passive avoidance paradigm under hypoxic stress. Hypoxia. 2018;6:35–56.
Wang Y, Chen Y, Wang C, Yang M, Wang Y, Bao L, et al. MIF is a 3’ flap nuclease that facilitates DNA replication and promotes tumor growth. Nat Commun. 2021;12:2954.
Huang W, Liu X, Zhang Y, Deng M, Li G, Chen G, et al. USP5 promotes breast cancer cell proliferation and metastasis by stabilizing HIF2alpha. J Cell Physiol. 2022;237:2211–9.
Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, et al. Parkin targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun. 2017;8:1823.
Ma B, Chen Y, Chen L, Cheng H, Mu C, Li J, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol. 2015;17:95–103.
Wang Y, Chen Y, Bao L, Zhang B, Wang JE, Kumar A, et al. CHD4 promotes breast cancer progression as a coactivator of hypoxia-inducible factors. Cancer Res. 2020;80:3880–91.
Acknowledgements
We thank The First Affiliated Hospital of Jinan University (JNU) and JNU Animal Studies Core for excellent help. We are grateful to Tongzheng Liu (Jinan University) for VHL and Cul2 plasmids. This work was supported by grants from the National Natural Science Foundation of China (81902691 and 82203297), Taishan Scholar Young Expert Program of Shandong Province (tsqn202306154 and tsqn202306155), Shandong Excellent Young Scientists Fund Program (Overseas) (2022HWYQ-077), Shandong Provincial Natural Science Foundation (ZR2023MH252), Guangdong Basic and Applied Basic Research Foundation (2022A1515011738 and 2021A1515011224), Guangzhou Science and Technology Program (202102080147).
Author information
Authors and Affiliations
Contributions
YC supervised the project; YC, Yijie Wang, and XL designed experiments. Yijie Wang, XL, MW, Yu Wang, SW, and LJ performed experiments. Yijie Wang, XL, ML, JZ, and YC analyzed the data. Yijie Wang, XL, ML, JZ, and YC wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Liu, X., Wang, M. et al. UBE3B promotes breast cancer progression by antagonizing HIF-2α degradation. Oncogene 42, 3394–3406 (2023). https://doi.org/10.1038/s41388-023-02842-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02842-z