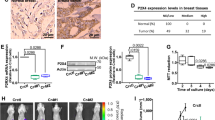
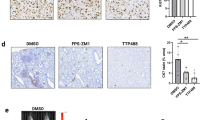
Abstract
Epithelial/Mesenchymal (E/M) plasticity plays a fundamental role both in embryogenesis and during tumorigenesis. The receptor for advanced glycation end products (RAGE) is a driver of cell plasticity in fibrotic diseases; however, its role and molecular mechanism in triple-negative breast cancer (TNBC) remains unclear. Here, we demonstrate that RAGE signaling maintains the mesenchymal phenotype of aggressive TNBC cells by enforcing the expression of SNAIL1. Besides, we uncover a crosstalk mechanism between the TGF-β and RAGE pathways that is required for the acquisition of mesenchymal traits in TNBC cells. Consistently, RAGE inhibition elicits epithelial features that block migration and invasion capacities. Next, since RAGE is a sensor of the tumor microenvironment, we modeled acute acidosis in TNBC cells and showed it promotes enhanced production of RAGE ligands and the activation of RAGE-dependent invasive properties. Furthermore, acute acidosis increases SNAIL1 levels and tumor cell invasion in a RAGE-dependent manner. Finally, we demonstrate that in vivo inhibition of RAGE reduces metastasis incidence and expands survival, consistent with molecular effects that support the relevance of RAGE signaling in E/M plasticity. These results uncover new molecular insights on the regulation of E/M phenotypes in cancer metastasis and provide rationale for pharmacological intervention of this signaling axis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq data were deposited in the Gene Expression Omnibus with the accession code GSE223229. The mass spectrometry proteomics data was deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD042042. All other data generated and backing the results and conclusions of this study are available from the corresponding author on reasonable request.
References
Foulkes W, Smith I, Reis-Filho J. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48.
Cardoso F. Global Status Metastatic Breast Cancer Report (2005–2015). Glob status metastatic breast cancer rep. 2016. http://www.breastcancervision.com.
Waks AG, Winer EP, Winer M. Breast cancer treatment: a review. JAMA. 2019;321:288–300.
Núñez Abad M, Calabuig-Fariñas S, Lobo de Mena M, José Godes Sanz de Bremond M, García González C, Torres, et al. Update on systemic treatment in early triple negative breast cancer. Ther Adv Med Oncol. 2021;13:1–18. https://doi.org/10.1177/1758835920986749.
Schmid P, Cortes J, Pusztai L, McArthur H, Kümmel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382:810–21. https://www.nejm.org/doi/full/10.1056/NEJMoa1910549.
Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–28.
Schmid P, Abraham J, Chan S, Wheatley D, Brunt AM, Nemsadze G, et al. Capivasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer: the PAKT trial. J Clin Oncol. 2020;423–33. https://pubmed.ncbi.nlm.nih.gov/31841354/.
Mendez O, Peg V, Salvans C, Pujals M, Fernandez Y, Abasolo I, et al. Extracellular HMGA1 promotes tumor invasion and metastasis in triple-negative breast cancer. Clin Cancer Res. 2018;24:6367–82.
Neeper M, Schmidt AM, Brett J, Shi Du Yan, Wang F, Pan YCE, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–5004. https://pubmed.ncbi.nlm.nih.gov/1378843/.
Kumar Pasupulati A, Chitra PS, Reddy GB. Advanced glycation end products mediated cellular and molecular events in the pathology of diabetic nephropathy. Biomol Concepts. 2016;7:293–9. https://pubmed.ncbi.nlm.nih.gov/27816946/.
Bongarzone S, Savickas V, Luzi F, Gee AD. Targeting the receptor for advanced glycation endproducts (RAGE): a medicinal chemistry perspective. J Med Chem. 2017;60:7213–32.
Narumi K, Miyakawa R, Ueda R, Hashimoto H, Yamamoto Y, Yoshida T, et al. Proinflammatory proteins S100A8/S100A9 activate NK cells via interaction with RAGE. J Immunol. 2015;194:5539–48. https://pubmed.ncbi.nlm.nih.gov/25911757/.
Hori O, Brett J, Slattery T, Cao R, Zhang J, Jing XC, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of RAGE and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–61. https://doi.org/10.1074/jbc.270.43.25752.
Ko S-Y, Ko H-A, Shieh T-M, Chang W-C, Chen H-I, Chang S-S, et al. Cell migration is regulated by AGE-RAGE interaction in human oral cancer cells in vitro. PLoS One. 2014;9:e110542 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4199749&tool=pmcentrez&rendertype=abstract.
Deane R, Yan SD, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, et al. RAGE mediates amyloid-β peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9:907–13.
Xie J, Méndez JD, Méndez-Valenzuela V, Aguilar-Hernández MM. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell Signal. 2013;25:2185–97. https://pubmed.ncbi.nlm.nih.gov/23838007/.
Yan SF, Ramasamy R, Schmidt AM. The RAGE axis a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res. 2010;106:842–53.
Cai Z, Liu N, Wang C, Qin B, Zhou Y, Xiao M, et al. Role of RAGE in Alzheimer’s disease. Cell Mol Neurobiol. 2016;36:483–95.
Ge X, Arriazu E, Magdaleno F, Antoine DJ, dela Cruz R, Theise N, et al. High mobility group box-1 drives fibrosis progression signaling via the receptor for advanced glycation end products in mice. Hepatology. 2018;68:2380–404. https://pubmed.ncbi.nlm.nih.gov/29774570/.
Xia JR, Liu NF, Zhu NX. Specific siRNA targeting the receptor for advanced glycation end products inhibits experimental hepatic fibrosis in rats. Int J Mol Sci. 2008;9:638–61. https://pubmed.ncbi.nlm.nih.gov/19325776/.
Riehl A, Németh J, Angel P, Hess J. The receptor RAGE: bridging inflammation and cancer. Cell Commun Signal. 2009;7:1–7. https://pubmed.ncbi.nlm.nih.gov/19426472/.
Nankali M, Karimi J, Goodarzi MT, Saidijam M, Khodadadi I, Razavi ANE, et al. Increased expression of the receptor for advanced glycation end-products (RAGE) is associated with advanced breast cancer stage. Oncol Res Treat. 2016;39:622–8.
Tesařová P, Kalousová M, Jáchymová M, Mestek O, Petruzelka L, Zima T. Receptor for advanced glycation end products (RAGE) - Soluble form (sRAGE) and gene polymorphisms in patients with breast cancer. Cancer Invest. 2007;25:720–5.
Jing R, Cui M, Wang J, Wang H. Receptor for advanced glycation end products (RAGE) soluble form (sRAGE): a new biomarker for lung cancer. Neoplasma. 2010;57:55–61. https://pubmed.ncbi.nlm.nih.gov/19895173/.
Li T, Qin W, Liu Y, Li S, Qin X, Liu Z. Effect of RAGE gene polymorphisms and circulating sRAGE levels on susceptibility to gastric cancer: a case-control study. Cancer Cell Int. 2017;17:1–10. https://cancerci.biomedcentral.com/articles/10.1186/s12935-017-0391-0.
Chen MC, Chen KC, Chang GC, Lin H, Wu CC, Kao WH, et al. RAGE acts as an oncogenic role and promotes the metastasis of human lung cancer. Cell Death Dis. 2020;11:1–13. https://doi.org/10.1038/s41419-020-2432-1.
Kwak T, Drews-Elger K, Ergonul A, Miller PC, Braley A, Hwang GH, et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene. 2017;36:1559–72. https://doi.org/10.1038/onc.2016.324.
Tian T, Li X, Hua Z, Ma J, Wu X, Liu Z, et al. S100A7 promotes the migration, invasion and metastasis of human cervical cancer cells through epithelial-mesenchymal transition. Oncotarget. 2017;8:24964–77. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5421902.
Palanissami G, Paul SFD. RAGE and its ligands: molecular interplay between glycation, inflammation, and hallmarks of cancer—a review. Horm Cancer. 2018;9:295–325. https://doi.org/10.1007/s12672-018-0342-9.
Weed SA, Parsons JT. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 2001;20:6418–34. https://pubmed.ncbi.nlm.nih.gov/11607842/.
Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–85. https://pubmed.ncbi.nlm.nih.gov/16051170/.
Weed SA, Du Y, Thomas Parsons J. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J Cell Sci. 1998;111:2433–43. https://pubmed.ncbi.nlm.nih.gov/9683637/.
Wang W, Liu Y, Liao K. Tyrosine phosphorylation of cortactin by the FAK-Src complex at focal adhesions regulates cell motility. BMC Cell Biol. 2011;12. https://pubmed.ncbi.nlm.nih.gov/22078467/.
Deane R, Singh I, Sagare AP, Bell RD, Ross NT, LaRue B, et al. A multimodal RAGE-specific inhibitor reduces amyloid β-mediated brain disorder in a mouse model of Alzheimer disease. J Clin Invest. 2012;122:1377–92.
Sabbagh MN, Agro A, Bell J, Aisen PS, Schweizer E, Galasko D. PF-04494700, an oral inhibitor of receptor for advanced glycation end products (RAGE), in Alzheimer disease. Alzheimer Dis Assoc Disord. 2011;25:206–12. https://pubmed.ncbi.nlm.nih.gov/21192237/.
Chen CR, Kang Y, Massagué J. Defective repression of c-myc in breast cancer cells: a loss at the core of the transforming growth factor beta growth arrest program. Proc Natl Acad Sci USA. 2001;98:992–9. https://pubmed.ncbi.nlm.nih.gov/11158583/.
Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43. https://pubmed.ncbi.nlm.nih.gov/15155580/.
Mejlvang J, Kriajevska M, Vandewalle C, Chernova T, Sayan AE, Berx G, et al. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol Biol Cell. 2007;18:4615–24. https://pubmed.ncbi.nlm.nih.gov/17855508/.
Cheng M, Liu H, Zhang D, Liu Y, Wang C, Liu F, et al. HMGB1 enhances the AGE-induced expression of CTGF and TGF-β via RAGE-dependent signaling in renal tubular epithelial cells. Am J Nephrol. 2015;41:257–66.
Serban AI, Stanca L, Geicu OI, Munteanu MC, Dinischiotu A. RAGE and TGF-β1 cross-talk regulate extracellular matrix turnover and cytokine synthesis in AGEs exposed fibroblast cells. PLoS One. 2016;11:e0152376.
Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ. The Receptor for Advanced Glycation End-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Signal. 2011;15:2175–84. https://pubmed.ncbi.nlm.nih.gov/21126167/.
Ray R, Jangde N, Singh SK, Sinha S, Rai V. Lysophosphatidic acid-RAGE axis promotes lung and mammary oncogenesis via protein kinase B and regulating tumor microenvironment. Cell Commun Signal. 2020;18. https://pubmed.ncbi.nlm.nih.gov/33109194/.
Ma H, Li SY, Xu P, Babcock SA, Dolence EK, Brownlee M, et al. Advanced glycation endproduct (AGE) accumulation and AGE receptor (RAGE) up-regulation contribute to the onset of diabetic cardiomyopathy. J Cell Mol Med. 2009;13:1751–64. https://pubmed.ncbi.nlm.nih.gov/19602045/.
Bai W, Zhou J, Zhou N, Liu Q, Cui J, Zou W, et al. Hypoxia-increased RAGE expression regulates chemotaxis and pro-inflammatory cytokines release through nuclear translocation of NF-κ B and HIF1α in THP-1 cells. Biochem Biophys Res Commun. 2018;495:2282–8. https://pubmed.ncbi.nlm.nih.gov/29258824/.
Rohani N, Hao L, Alexis MS, Joughin BA, Krismer K, Moufarrej MN, et al. Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 2019;79:1952–66.
Yatime L, Andersen GR. Structural insights into the oligomerization mode of the human receptor for advanced glycation end-products. FEBS J. 2013;280:6556–68.
Jangde N, Ray R, Rai V. RAGE and its ligands: from pathogenesis to therapeutics. Crit Rev Biochem Mol Biol. 2020;55:555–75. https://doi.org/10.1080/10409238.2020.1819194.
Hudson BI, Lippman ME. Targeting RAGE signaling in inflammatory disease. Annu Rev Med. 2018;69:24.1–24.16. https://pubmed.ncbi.nlm.nih.gov/29106804/.
Wang L, Wang HL, Liu TT, Lan HY. TGF-beta as a master regulator of diabetic nephropathy. Int J Mol Sci. 2021;22:1–18. https://doi.org/10.3390/ijms22157881.
Raghavan CT, Nagaraj RH. AGE-RAGE interaction in the TGFβ2-mediated epithelial to mesenchymal transition of human lens epithelial cells. Glycoconj J. 2016;33:631–43. https://doi.org/10.1007/s10719-016-9686-y.
Nam MH, Pantcheva MB, Rankenberg J, Nagaraj RH. Transforming growth factor-β2-mediated mesenchymal transition in lens epithelial cells is repressed in the absence of RAGE. Biochem J. 2021;478:2285–96.
Chen YC, Statt S, Wu R, Chang HT, Liao JW, Wang CN, et al. High mobility group box 1-induced epithelial mesenchymal transition in human airway epithelial cells. Sci Rep. 2016;6:18815.
He M, Kubo H, Ishizawa K, Hegab AE, Yamamoto Y, Yamamoto H, et al. The role of the receptor for advanced glycation end-products in lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2007;293:1427–36.
Yin C, Li H, Zhang B, Liu Y, Lu G, Lu S, et al. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial-mesenchymal transition. Breast Cancer Res Treat. 2013;142:297–309.
Zhang J, Shao S, Han D, Xu Y, Jiao D, Wu J, et al. High mobility group box 1 promotes the epithelial-to-mesenchymal transition in prostate cancer PC3 cells via the RAGE/NF-KB signaling pathway. Int J Oncol. 2018;53:659–71.
Chen S, Jim B, Ziyadeh FN. Diabetic nephropathy and transforming growth factor-β: transforming our view of glomerulosclerosis and fibrosis build-up. Semin Nephrol. 2003;23:532–43.
Hu HH, Chen DQ, Wang YN, Feng YL, Cao G, Vaziri ND, et al. New insights into TGF-β/Smad signaling in tissue fibrosis. Chem Biol Interact. 2018;292:76–83.
Li JH, Huang XR, Zhu HJ, Oldfield M, Cooper M, Truong LD, et al. Advanced glycation end products activate Smad signaling via TGF-beta-dependent and independent mechanisms: implications for diabetic renal and vascular disease. FASEB J. 2004;18:176–8. https://pubmed.ncbi.nlm.nih.gov/12709399/.
Chung ACK, Zhang H, Kong YZ, Tan JJ, Huang XR, Kopp JB, et al. Advanced glycation end-products induce tubular CTGF via TGF-β-independent Smad3 signaling. J Am Soc Nephrol. 2010;21:249–60.
Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Activation of the Erk pathway is required for TGF-b1 – induced EMT in vitro. Neoplasia. 2004;6:603–10.
Li Y, Wang P, Zhao J, Li H, Liu D, Zhu W. HMGB1 attenuates TGF-β-induced epithelial–mesenchymal transition of FaDu hypopharyngeal carcinoma cells through regulation of RAGE expression. Mol Cell Biochem. 2017;431:1–10.
Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. 2020;5. https://doi.org/10.1038/s41392-020-00280-x.
Corbet C, Feron O. Tumour acidosis: from the passenger to the driver’s seat. Nat Rev Cancer. 2017;17:577–93.
Sadeghi M, Ordway B, Rafiei I, Borad P, Fang B, Koomen JL, et al. Integrative analysis of breast cancer cells reveals an epithelial-mesenchymal transition role in adaptation to acidic microenvironment. Front Oncol. 2020;10:1–14.
Riemann A, Schneider B, Ihling A, Nowak M, Sauvant C, Thews O, et al. Acidic environment leads to ROS-Induced MAPK signaling in cancer cells. PLoS One. 2011;6:e22445.
Chen B, Liu J, Ho TT, Ding X, Mo YY. Erk-mediated nf-κb activation through asic1 in response to acidosis. Oncogenesis. 2016;5:1–8.
Sauvant C, Nowak M, Wirth C, Schneider B, Riemann A, Gekle M, et al. Acidosis induces multi-drug resistance in rat prostate cancer cells (AT1) in vitro and in vivo by increasing the activity of the p-glycoprotein via activation of p38. Int J Cancer. 2008;123:2532–42. https://onlinelibrary.wiley.com/doi/full/10.1002/ijc.23818.
Gupta SC, Singh R, Pochampally R, Watabe K, Mo Y-Y. Acidosis promotes invasiveness of breast cancer cells through ROS-AKT-NF-kB pathway. Oncotarget. 2014;5:12070–82. http://www.oncotarget.com/fulltext/2514.
Suzuki A, Maeda T, Baba Y, Shimamura K, Kato Y. Acidic extracellular ph promotes epithelial mesenchymal transition in lewis lung carcinoma model. Cancer Cell Int. 2014;14:1–11.
Peppicelli S, Bianchini F, Torre E, Calorini L. Contribution of acidic melanoma cells undergoing epithelial-to-mesenchymal transition to aggressiveness of non-acidic melanoma cells. Clin Exp Metastasis. 2014;31:423–33. https://pubmed.ncbi.nlm.nih.gov/24469963/.
Xie J, Reverdatto S, Frolov A, Hoffmann R, Burz DS, Shekhtman A. Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE). J Biol Chem. 2008;283:27255–69. https://pubmed.ncbi.nlm.nih.gov/18667420/.
Zong H, Madden A, Ward M, Mooney M, Elliott C, Stitt A. Homodimerization is essential for the receptor for advanced glycation end products (RAGE)-mediated signal transduction. J Biol Chem. 2010;285:23137–46. https://pubmed.ncbi.nlm.nih.gov/20504772/.
Gebhardt C, Riehl A, Durchdewald M, Németh J, Fürstenberger G, Müller-Decker K, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. 2008;205:275–85.
Kang R, Tang D, Lotze MT, Zeh HJ. AGER/RAGE-mediated autophagy promotes pancreatic tumorigenesis and bioenergetics through the IL6-pSTAT3 pathway. Autophagy. 2012;8:989–91.
Burstein AH, Grimes I, Galasko DR, Aisen PS, Sabbagh M, Mjalli AMM. Effect of TTP488 in patients with mild to moderate Alzheimer’s disease. BMC Neurol. 2014;14:1–8. https://pubmed.ncbi.nlm.nih.gov/24423155/.
Galasko D, Bell J, Mancuso JY, Kupiec JW, Sabbagh MN, Van Dyck C, et al. Clinical trial of an inhibitor of RAGE-Ab interactions in Alzheimer disease. Neurology. 2014;82:1536–42.
ClinicalTrials.gov. Evaluation of the efficacy and safety of azeliragon (TTP488) in patients with mild Alzheimer’s disease (STEADFAST). Identifier (NCT number): NCT02080364. 2014.
Gregori J, Villarreal L, Méndez O, Sánchez A, Baselga J, Villanueva J. Batch effects correction improves the sensitivity of significance tests in spectral counting-based comparative discovery proteomics. J Proteom. 2012;75:3938–51. https://www.sciencedirect.com/science/article/pii/S1874391912002758.
Bellio C, Emperador M, Castellano P, Gris-Oliver A, Canals F, Sánchez-Pla A, et al. GDF15 is an eribulin response biomarker also required for survival of DTP breast cancer cells. Cancers. 2022;14:1–23.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. 2010. http://genomebiology.com/2010/11/3/R25.
Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. http://bioconductor.org.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. https://academic.oup.com/nar/article/43/7/e47/2414268.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://pubmed.ncbi.nlm.nih.gov/16199517/.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–7.
Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. https://pubmed.ncbi.nlm.nih.gov/1172191/.
Funding
Authors from VHIO would like to acknowledge the Cellex Foundation for providing research facilities and equipment, the FERO Foundation for their funding support and the Centro de Investigación Biomédica en Red de Cáncer (CIBERONC) from the Institute of Health Carlos III (ISCIII) for their support on this research. JV acknowledges the Project “PI19/01292", funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union. Authors from VHIO acknowledge the State Agency for Research (Agencia Estatal de Investigación) the financial support as a Center of Excellence Severo Ochoa (CEX2020-001024-S/AEI/10.13039/ 501100011033).
Author information
Authors and Affiliations
Contributions
Conceptualization, MP and JV; Funding acquisition, JT and JV; Investigation, MP, CM, CB and OM; Methodology, MP, CM, EG, MA, RF and FC; Project administration, JV; Resources, EZ, CS, LP, PN and FM; Bioinformatic Analysis, MA-C and LN; Supervision, JV; Validation, MP, CM and JV; Writing – original draft, MP and JV; Writing – review & editing, MP, CM and JV.
Corresponding author
Ethics declarations
Competing interests
JT reports personal financial interest in form of scientific consultancy role for Array Biopharma, AstraZeneca, Bayer, Boehringer Ingelheim, Cardiff Oncology, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche Ltd, Genentech Inc, HalioDX SAS, Hutchison MediPharma International, Ikena Oncology, Inspirna Inc, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Scandion Oncology, Scorpion Therapeutics, Seattle Genetics, Servier, Sotio Biotech, Taiho, Tessa Therapeutics, TheraMyc and Tolremo Therapeutics. Stocks: Oniria Therapeutics and also educational collaboration with Imedex/HMP, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education and Physicians Education Resource (PER). PN reports personal financial interest in form of receiving honoraria or consultation fees by Novartis, Bayer, MSD Oncology, Targos Molecular Pathology GmbH. Travel, accommodation paid or reimbursed by Novartis. CS reports personal financial interest serving as consultant, participated in advisory boards or received travel grants from: AstraZeneca, AX'Consulting, Byondis B.V, Daiichi Sankyo, Eisai, Exact Sciences, Exeter Pharma, F.Hoffmann-La Roche Ltd, Gilead, Lilly, MediTech, Merck Sharp & Dohme, Novartis, Pfizer, Philips, Pierre Fabre, PintPharma, Puma Biotechnology, SeaGen, Synthon biopharmaceutical and Zymeworks. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pujals, M., Mayans, C., Bellio, C. et al. RAGE/SNAIL1 signaling drives epithelial-mesenchymal plasticity in metastatic triple-negative breast cancer. Oncogene 42, 2610–2628 (2023). https://doi.org/10.1038/s41388-023-02778-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02778-4