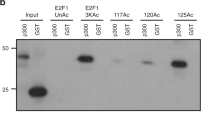
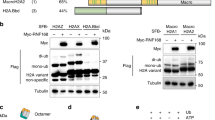
Abstract
Ewing sarcoma breakpoint region 1 (EWSR1) is a member of FET (FUS/EWSR1/TAF15) RNA-binding family of proteins. The Ewing sarcoma oncoprotein EWS-FLI1 has been extensively studied, while much less is known about EWSR1 itself, especially the potential role of EWSR1 in response to DNA damage. Here, we found that UV irradiation induces acetylation of EWSR1, which is required for its nucleoli translocation. We identified K423, K432, K438, K640, and K643 as the major acetylation sites, p300/CBP and HDAC3/HDAC10 as the major acetyltransferases and deacetylases, respectively. Mechanically, UV-induced EWSR1 acetylation repressed its interaction with spliceosomal component U1C, which caused abnormal splicing of CHK2, suppressing the activity of CHK2 in response to UV irradiation. Taken together, our findings uncover acetylation as a novel regulatory modification of EWSR1, and is essential for its function in DNA damage response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Prim. 2018;4:5.
Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992;359:162–5.
Alex D, Lee KA. RGG-boxes of the EWS oncoprotein repress a range of transcriptional activation domains. Nucleic Acids Res. 2005;33:1323–31.
Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18:1489–97.
Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Investig. 2007;117:1314–23.
Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45.
Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85.
Hardin JD, Boast S, Schwartzberg PL, Lee G, Alt FW, Stall AM, et al. Bone marrow B lymphocyte development in c-abl-deficient mice. Cell Immunol. 1995;165:44–54.
Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–71.
Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–50.
Lee SG, Kim N, Kim SM, Park IB, Kim H, Kim S, et al. Ewing sarcoma protein promotes dissociation of poly(ADP-ribose) polymerase 1 from chromatin. EMBO Rep. 2020;21:e48676.
Spahn L, Petermann R, Siligan C, Schmid JA, Aryee DN, Kovar H. Interaction of the EWS NH2 terminus with BARD1 links the Ewing’s sarcoma gene to a common tumor suppressor pathway. Cancer Res. 2002;62:4583–7.
Paronetto MP, Miñana B, Valcárcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43:353–68.
Mustofa MK, Tanoue Y, Tateishi C, Vaziri C, Tateishi S. Roles of Chk2/CHEK2 in guarding against environmentally induced DNA damage and replication-stress. Environ Mol Mutagen. 2020;61:730–5.
Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6:442–57.
Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–19.
Da Costa IC, Schmidt CK. Ubiquitin-like proteins in the DNA damage response: the next generation. Essays Biochem. 2020;64:737–52.
Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33.
Deloulme JC, Prichard L, Delattre O, Storm DR. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J Biol Chem. 1997;272:27369–77.
Kim J, Lee JM, Branton PE, Pelletier J. Modification of EWS/WT1 functional properties by phosphorylation. Proc Natl Acad Sci USA. 1999;96:14300–5.
Kim J, Lee JM, Branton PE, Pelletier J. Modulation of EWS/WT1 activity by the v-Src protein tyrosine kinase. FEBS Lett. 2000;474:121–8.
Guinamard R, Fougereau M, Seckinger P. The SH3 domain of Bruton’s tyrosine kinase interacts with Vav, Sam68 and EWS. Scand J Immunol. 1997;45:587–95.
Klevernic IV, Morton S, Davis RJ, Cohen P. Phosphorylation of Ewing’s sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochem J. 2009;418:625–34.
Araya N, Hiraga H, Kako K, Arao Y, Kato S, Fukamizu A. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem Biophys Res Commun. 2005;329:653–60.
Kim JD, Kako K, Kakiuchi M, Park GG, Fukamizu A. EWS is a substrate of type I protein arginine methyltransferase, PRMT8. Int J Mol Med. 2008;22:309–15.
Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011;278:988–98.
Bachmaier R, Aryee DN, Jug G, Kauer M, Kreppel M, Lee KA, et al. O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing’s sarcoma. Oncogene. 2009;28:1280–4.
Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21.
Paronetto MP. Ewing sarcoma protein: a key player in human cancer. Int J Cell Biol. 2013;2013:642853.
Muto Y, Pomeranz Krummel D, Oubridge C, Hernandez H, Robinson CV, Neuhaus D, et al. The structure and biochemical properties of the human spliceosomal protein U1C. J Mol Biol. 2004;341:185–98.
Knoop LL, Baker SJ. The splicing factor U1C represses EWS/FLI-mediated transactivation. J Biol Chem. 2000;275:24865–71.
Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5’ splice site in the absence of base pairing. Nature. 2002;419:86–90.
Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512.
Tanoue Y, Toyoda T, Sun J, Mustofa MK, Tateishi C, Endo S, et al. Differential roles of Rad18 and Chk2 in genome maintenance and skin carcinogenesis following UV exposure. J Investig Dermatol. 2018;138:2550–7.
Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:456–62.
Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–9.
Dutto I, Scalera C, Prosperi E. CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell Mol Life Sci. 2018;75:1325–38.
Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–40.
Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–8.
Shandilya J, Swaminathan V, Gadad SS, Choudhari R, Kodaganur GS, Kundu TK. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol Cell Biol. 2009;29:5115–27.
Das S, Cong R, Shandilya J, Senapati P, Moindrot B, Monier K, et al. Characterization of nucleolin K88 acetylation defines a new pool of nucleolin colocalizing with pre-mRNA splicing factors. FEBS Lett. 2013;587:417–24.
Muñoz MJ, Pérez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–20.
Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, et al. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–66.
Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–7.
Berge EO, Staalesen V, Straume AH, Lillehaug JR, Lønning PE. Chk2 splice variants express a dominant-negative effect on the wild-type Chk2 kinase activity. Biochim Biophys Acta. 2010;1803:386–95.
Staalesen V, Falck J, Geisler S, Bartkova J, Børresen-Dale AL, Lukas J, et al. Alternative splicing and mutation status of CHEK2 in stage III breast cancer. Oncogene. 2004;23:8535–44.
Ren X, Long M, Li Z, Wu B, Jin T, Tu C, et al. Oncogene PRR14 promotes breast cancer through activation of PI3K signal pathway and inhibition of CHEK2 pathway. Cell Death Dis. 2020;11:464.
Srivastava A, Giangiobbe S, Skopelitou D, Miao B, Paramasivam N, Diquigiovanni C, et al. Whole genome sequencing prioritizes CHEK2, EWSR1, and TIAM1 as possible predisposition genes for familial non-medullary thyroid cancer. Front Endocrinol. 2021;12:600682.
Han C, Khodadadi-Jamayran A. SF3B1 homeostasis is critical for survival and therapeutic response in T cell leukemia. Sci Adv. 2022;8:eabj8357.
Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem. 2015;84:355–79.
Rösel TD, Hung LH, Medenbach J, Donde K, Starke S, Benes V, et al. RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J. 2011;30:1965–76.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (81874147, 82172959). We thank core facility at Peking University Health Science Center for experiment help.
Author information
Authors and Affiliations
Contributions
TZ and JL designed the research; TZ, ZW, ML, LL, XY, YZ, JB, and YL performed the experiments; TZ, ZW, MR, CS, and WW analyzed data; TZ, HT, JL drafted the manuscript; TZ and JL modified the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, T., Wang, Z., Liu, M. et al. Acetylation dependent translocation of EWSR1 regulates CHK2 alternative splicing in response to DNA damage. Oncogene 41, 3694–3704 (2022). https://doi.org/10.1038/s41388-022-02383-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02383-x
This article is cited by
-
The nexus between RNA-binding proteins and their effectors
Nature Reviews Genetics (2023)