Abstract
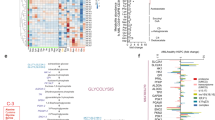
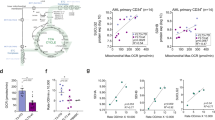
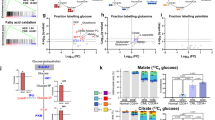
We characterize the metabolic background in distinct Acute Myeloid Leukemias (AMLs), by comparing the metabolism of primary AML blasts isolated at diagnosis with that of normal hematopoietic maturing progenitors, using the Seahorse XF Agilent. Leukemic cells feature lower spare respiratory (SRC) and glycolytic capacities as compared to hematopoietic precursors (i.e. day 7, promyelocytes). According with Proton Leak (PL) values, AML blasts can be grouped in two well defined populations. The AML group with blasts presenting high PL or high basal OXPHOS plus high SRC levels had shorter overall survival time and significantly overexpressed myeloid cell leukemia 1 (MCL1) protein. We demonstrate that MCL1 directly binds to Hexokinase 2 (HK2) on the outer mitochondrial membrane (OMM). Overall, these results suggest that high PL and high SRC plus high basal OXPHOS levels at disease onset, arguably with the concourse of MCL1/HK2 action, are significantly linked with shorter overall survival time in AML. Our data describe a new function for MCL1 protein in AMLs’ cells: by forming a complex with HK2, MCL1 co-localizes to VDAC on the OMM, thus inducing glycolysis and OXPHOS, ultimately conferring metabolic plasticity and promoting resistance to therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Cassim S, Vučetić M, Ždralević M, Pouyssegur J. Warburg and beyond: the power of mitochondrial metabolism to collaborate or replace fermentative glycolysis in cancer. Cancers. 2020;12:1119.
Samudio I, Fiegl M, McQueen T, Clise-Dwyer K, Andreeff M. The warburg effect in leukemia-stroma cocultures is mediated by mitochondrial uncoupling associated with uncoupling protein 2 activation. Cancer Res. 2008;68:5198–205.
Robinson AJ, Hopkins GL, Rastogi N, Hodges M, Doyle M, Davies S, et al. Reactive oxygen species drive proliferation in acute myeloid leukemia via the glycolytic regulator PFKFB3. Cancer Res. 2020;80:937–49.
Warr MR, Pietras EM, Passegué E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med. 2011;3:681–701.
Peng M, Huang Y, Zhang L, Zhao X, Hou Y. Targeting mitochondrial oxidative phosphorylation eradicates acute myeloid leukemic stem cells. Front Oncol. 2022;12:899502.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41.
Kühlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89.
Divakaruni AS, Brand MD. The regulation and physiology of mitochondrial proton leak. Physiology. 2011;26:192–205.
Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–56.
Lunt SY, Vander, Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–64.
Ortega AD, Sánchez-Aragó M, Giner-Sánchez D, Sánchez-Cenizo L, Willers I, Cuezva JM. Glucose avidity of carcinomas. Cancer Lett. 2009;276:125–35.
Chen Y, Liang Y, Luo X, Hu Q. Oxidative resistance of leukemic stem cells and oxidative damage to hematopoietic stem cells under pro-oxidative therapy. Cell Death Dis. 2020;11:291.
Noguera NI, Pelosi E, Angelini DF, Piredda ML, Guerrera G, Piras E, et al. High-dose ascorbate and arsenic trioxide selectively kill acute myeloid leukemia and acute promyelocytic leukemia blasts in vitro. Oncotarget. 2017;8:32550–65.
Nair RR, Tolentino JH, Hazlehurst LA. Role of STAT3 in transformation and drug resistance in CML. Front Oncol. 2012;2:30.
Chen Y, Li J, Zhao Z. Redox control in acute lymphoblastic leukemia: from physiology to pathology and therapeutic opportunities. Cells. 2021;10:1218.
Zhou Y, Guo Y, Tam KY. Targeting glucose metabolism to develop anticancer treatments and therapeutic patents. Expert Opin Ther Pat. 2022;32:441–53.
Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015;22:364.
Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–57.
Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–96.
John S, Weiss JN, Ribalet B. Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One. 2011;6:e17674.
da-Silva WS, Gómez-Puyou A, de Gómez-Puyou MT, Moreno-Sanchez R, De Felice FG, de Meis L, et al. Mitochondrial bound hexokinase activity as a preventive antioxidant defense: steady-state ADP formation as a regulatory mechanism of membrane potential and reactive oxygen species generation in mitochondria. J Biol Chem. 2004;279:39846–55.
Galluzzi L, Kepp O, Tajeddine N, Kroemer G. Disruption of the hexokinase-VDAC complex for tumor therapy. Oncogene. 2008;27:4633–5.
Thomas GE, Egan G, García-Prat L, Botham A, Voisin V, Patel PS, et al. The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness. Nat Cell Biol. 2022;24:872–84.
Banella C, Catalano G, Travaglini S, Pelosi E, Ottone T, Zaza A, et al. Ascorbate plus buformin in AML: a metabolic targeted treatment. Cancers. 2022;14:2565.
Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88.
Wang H, Guo M, Wei H, Chen Y. Targeting MCL-1 in cancer: current status and perspectives. J Hematol Oncol. 2021;14:67.
Glaser SP, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD, et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012;26:120–5.
Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, et al. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest. 2010;120:2109–18.
Carter BZ, Mak PY, Tao W, Warmoes M, Lorenzi PL, Mak D, et al. Targeting MCL-1 dysregulates cell metabolism and leukemia-stroma interactions and resensitizes acute myeloid leukemia to BCL-2 inhibition. Haematologica. 2020;12:23.
Rasmussen ML, Kline LA, Park KP, Ortolano NA, Romero-Morales AI, Anthony CC, et al. A non-apoptotic function of MCL-1 in promoting pluripotency and modulating mitochondrial dynamics in stem cells. Stem Cell Rep. 2018;10:684–92.
Jamil S, Sobouti R, Hojabrpour P, Raj M, Kast J, Duronio V. A proteolytic fragment of Mcl-1 exhibits nuclear localization and regulates cell growth by interaction with Cdk1. Biochem J. 2005;387:659–67.
Ariës IM, Hansen BR, Koch T, van den Dungen R, Evans WE, Pieters R, et al. The synergism of MCL1 and glycolysis on pediatric acute lymphoblastic leukemia cell survival and prednisolone resistance. Haematologica. 2013;98:1905–11.
Banella C, Ginevrino M, Catalano G, Fabiani E, Falconi G, Divona M, et al. Absence of FGFR3-TACC3 rearrangement in hematological malignancies with numerical chromosomal alteration. Hematol Oncol Stem Cell Ther. 2021;14:163–8.
Noguera NI, Piredda ML, Taulli R, Catalano G, Angelini G, Gaur G, et al. PML/RARa inhibits PTEN expression in hematopoietic cells by competing with PU.1 transcriptional activity. Oncotarget. 2016;7:66386–97.
Quattrocchi A, Maiorca C, Billi M, Tomassini S, De Marinis E, Cenfra N, et al. Genetic lesions disrupting calreticulin 3’-untranslated region in JAK2 mutation-negative polycythemia vera. Am J Hematol. 2020:E263–E267. https://doi.org/10.1002/ajh.25911.
Albanesi J, Noguera NI, Banella C, Colangelo T, De Marinis E, Leone S, et al. Transcriptional and metabolic dissection of ATRA-induced granulocytic differentiation in NB4 acute promyelocytic leukemia cells. Cells. 2020;9:2423.
Noguera NI, Song MS, Divona M, Catalano G, Calvo KL, García F, et al. Nucleophosmin/B26 regulates PTEN through interaction with HAUSP in acute myeloid leukemia. Leukemia. 2013;27:1037–43.
Piredda ML, Gaur G, Catalano G, Divona M, Banella C, Travaglini S, et al. PML/RARA inhibits expression of HSP90 and its target AKT. Br J Haematol. 2019;184:937–48.
Banella C, Catalano G, Travaglini S, Divona M, Masciarelli S, Guerrera G, et al. PML/RARa interferes with NRF2 transcriptional activity increasing the sensitivity to ascorbate of acute promyelocytic leukemia cells. Cancers. 2019;12:95.
Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–54.
Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125:2120–30.
Bi G, Bian Y, Liang J, Yin J, Li R, Zhao M, et al. Pan-cancer characterization of metabolism-related biomarkers identifies potential therapeutic targets. J Transl Med. 2021;19:219.
Lee KM, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–647.e637.
Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M, et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol. 2012;14:575–83.
Senichkin VV, Streletskaia AY, Gorbunova AS, Zhivotovsky B, Kopeina GS. Saga of Mcl-1: regulation from transcription to degradation. Cell Death Differ. 2020;27:405–19.
Sun L, Shukair S, Naik TJ, Moazed F, Ardehali H. Glucose phosphorylation and mitochondrial binding are required for the protective effects of hexokinases I and II. Mol Cell Biol. 2008;28:1007–17.
Chiara F, Castellaro D, Marin O, Petronilli V, Brusilow WS, Juhaszova M, et al. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS One. 2008;3:e1852.
Bagchi S, Fredriksson R, Wallén-Mackenzie Å. In situ proximity ligation assay (PLA). Methods Mol Biol. 2015;1318:149–59.
Kreitz J, Schönfeld C, Seibert M, Stolp V, Alshamleh I, Oellerich T, et al. Metabolic plasticity of acute myeloid leukemia. Cells. 2019;8:805.
Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547:104–8.
Marchetti P, Fovez Q, Germain N, Khamari R, Kluza J. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020;34:13106–24.
Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–54.
Nelson H, Coomber BL. Hexokinase II expression is correlated with colorectal cancer prognosis. Cancer Treat Commun. 2016;6:11–16.
Pedersen PL. Warburg, me and Hexokinase 2: multiple discoveries of key molecular events underlying one of cancers’ most common phenotypes, the “Warburg Effect”, i.e., elevated glycolysis in the presence of oxygen. J Bioenerg Biomembr. 2007;39:211–22.
Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277:7610–8.
Huang H, Shah K, Bradbury NA, Li C, White C. Mcl-1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis. 2014;5:e1482.
Rashkovan M, Ferrando A. Metabolic dependencies and vulnerabilities in leukemia. Genes Dev. 2019;33:1460–74.
Noguera NI, Hasan SK, Ammatuna E, Venditti A. Editorial: metabolic rewiring in leukemias. Front Oncol. 2021;11:775167.
Acknowledgements
This research was funded by AIRC 5 ×1000 call “Metastatic disease: the key unmet need in oncology” to MYNERVA project, #21267 (MYeloid NEoplasms Research Venture AIRC. A detailed description of the MYNERVA project is available at http://www.progettoagimm.it, accessed on 2 April 2022) to MTV, by PRIN 2017WXR7ZT_004 to MTV and by PRIN 2017WWB99Z to CN.
Author information
Authors and Affiliations
Contributions
GC co-wrote the manuscript, contributed to study design and analyzed the results; AZ, CB, ST and AS performed experiments; TO, MD, IP, FB and LM obtained and characterized patient samples, updated the clinical data; EP, GC and UT provided cord blood samples, carried out the differentiation experiments of CD34+ progenitors; EdM, EA, CN and AV analyzed the data and co-wrote the manuscript, NIN and MTV contributed to study design, analyzed the experiments and co-wrote the manuscript. All authors have read and agreed with the content of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Catalano, G., Zaza, A., Banella, C. et al. MCL1 regulates AML cells metabolism via direct interaction with HK2. Metabolic signature at onset predicts overall survival in AMLs’ patients. Leukemia 37, 1600–1610 (2023). https://doi.org/10.1038/s41375-023-01946-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01946-5
This article is cited by
-
Development and validation of a cuproptosis-related prognostic model for acute myeloid leukemia patients using machine learning with stacking
Scientific Reports (2024)
-
Abatement of the binding of human hexokinase II enzyme monomers by in-silico method with the design of inhibitory peptides
In Silico Pharmacology (2024)