Abstract
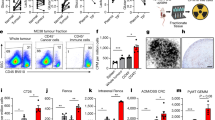
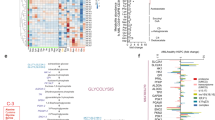
Acute myeloid leukaemia (AML) cells metabolise glucose by glycolysis-based re-programming. However, how glucose uptake is partitioned between leukaemia cells and other cells of the bone marrow micro-environment is unstudied. We used a positron emission tomography (PET) tracer 18F fluorodeoxyglucose ([18F]-FDG) probe and transcriptomic analyses to detect glucose uptake by diverse cells in the bone marrow micro-environment in a MLL-AF9-induced mouse model. Leukaemia cells had the greatest glucose uptake with leukaemia stem and progenitor cells having the greatest glucose uptake. We also show the effects of anti-leukaemia drugs on leukaemia cell numbers and glucose uptake. Our data suggest targeting glucose uptake as a potential therapy strategy in AML if our observations are validated in humans with AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data are available in the text and supplement. Raw transcriptomic data have been submitted to GEO database under the accession number GSE227584. R scripts are available upon reasonable request from HZ.
Code availability
Data are available in the text and supplement. Raw transcriptomic data have been submitted to GEO database under the accession number GSE227584. R scripts are available upon reasonable request from HZ.
References
Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–69.
Jones CL, Stevens BM, Pollyea DA, Culp-Hill R, Reisz JA, Nemkov T, et al. Nicotinamide metabolism mediates resistance to venetoclax in relapsed acute myeloid leukemia stem cells. Cell Stem Cell. 2020;27:748–64.
Stevens BM, Jones CL, Pollyea DA, Culp-Hill R, D’Alessandro A, Winters A, et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat Cancer. 2020;1:1176–87.
Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T. et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2019;35:333–5.
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20.
Ye H, Adane B, Khan N, Alexeev E, Nusbacher N, Minhajuddin M, et al. Subversion of systemic glucose metabolism as a mechanism to support the growth of leukemia cells. Cancer Cell. 2018;34:659–73.
Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593:282–8.
Saito Y, Chapple RH, Lin A, Kitano A, Nakada D. AMPK protects leukemia-initiating cells in myeloid leukemias from metabolic stress in the bone marrow. Cell Stem Cell. 2015;17:585–96.
Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–49.
Zhang Y, Wang D, Peng M, Tang L, Ouyang J, Xiong F, et al. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res. 2021;40:81.
Eberwine J, Sul J-Y, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11:25–7.
Huang D, Sun G, Hao X, He X, Zheng Z, Chen C, et al. ANGPTL2-containing small extracellular vesicles from vascular endothelial cells accelerate leukemia progression. J Clin Invest. 2021;131:e138986.
Gao A, Gong Y, Zhu C, Yang W, Li Q, Zhao M, et al. Bone marrow endothelial cell-derived interleukin-4 contributes to thrombocytopenia in acute myeloid leukemia. Haematologica. 2019;104:1950–61.
Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJ, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27:852–60.
Hassanein M, Hight MR, Buck JR, Tantawy MN, Nickels ML, Hoeksema MD, et al. Preclinical evaluation of 4-[18F]Fluoroglutamine PET to assess ASCT2 expression in lung cancer. Mol Imaging Biol. 2016;18:18–23.
Stölzel F, Lüer T, Löck S, Parmentier S, Kuithan F, Kramer M, et al. The prevalence of extramedullary acute myeloid leukemia detected by FDG-PET/CT: final results from the prospective PETAML trial. Haematologica. 2020;105:1552–8.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902.e21.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
de Beauchamp L, Himonas E, Helgason GV. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2022;36:1–12.
Joshi SK, Nechiporuk T, Bottomly D, Piehowski PD, Reisz JA, Pittsenbarger J, et al. The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell. 2021;39:999–1014.
Schuurhuis GJ, Heuser M, Freeman S, Béné M-C, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131:1275–91.
Ravandi F, Walter RB, Freeman SD. Evaluating measurable residual disease in acute myeloid leukemia. Blood Adv. 2018;2:1356–66.
Acknowledgements
Drs. Xiaoping Yi (Department of Radiology, Xiangya Hospital, Central South University, Changsha, Hunan, China) and Xiaoxia Hu (Collaborative Innovation Centre of Haematology, Shanghai Jiao Tong University School of Medicine, Shanghai, China) kindly reviewed the typescript. RPG acknowledges support from the National Institute of Health Research (NIHR) Biomedical Research Centre.
Funding
Supported, in part, by the Talent Young Programme of Guangdong Province (2021B1515020017), National Natural Science Foundation of China (Grant No. 81970143 and No. 82270167), Municipal School Joint Programme from Guangzhou Science and Technological Project (202201020012) and the Leading Talents Programme from The First Affiliated Hospital of Jinan University to HZ.
Author information
Authors and Affiliations
Contributions
SQD data curation, experiment conduction, statistical and transcriptomic analysis, visualisation, preparing the typescript. JD data curation, result interpretation, investigation, preparing and editing the typescript. RPG, data interpretation, reviewing and editing the typescript. LW technical support. HEZ, FSL, KXH experimental assistance. HX technical support. HZ project design, investigation, data interpretation, reviewing and editing the typescript. All authors take responsibility for the content of the typescript and agreed to submit for publication.
Corresponding author
Ethics declarations
Competing interests
RPG is a consultant to NexImmune Inc. Nanexa Pharma Ascentage Pharm Group and Antengene Biotech LLC, Medical Director of FFF Enterprises Inc.; Partner in AZCA Inc.; Board of Directors of Russian Foundation for Cancer Research Support and Scientific Advisory Board: StemRad Ltd.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, S., Du, J., Gale, R.P. et al. Glucose partitioning in the bone marrow micro-environment in acute myeloid leukaemia. Leukemia 37, 1407–1412 (2023). https://doi.org/10.1038/s41375-023-01912-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01912-1