Abstract
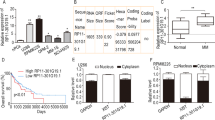
The biological role and therapeutic potential of long non-coding RNAs (lncRNAs) in multiple myeloma (MM) are still open questions. Herein, we investigated the functional significance of the oncogenic lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) in MM. Our study demonstrates that NEAT1 expression level is higher in MM than in the majority of hematological malignancies. NEAT1 silencing by novel LNA-gapmeR antisense oligonucleotide inhibits MM cell proliferation and triggers apoptosis in vitro and in vivo murine MM model as well. By transcriptome analyses, we found that NEAT1 targeting downregulates genes involved in DNA repair processes including the Homologous Recombination pathway, which in turn results in massive DNA damage. These findings may explain the synergistic impact on apoptosis observed in MM cell lines co-treated with inhibitors of both NEAT1 and PARP. The translational significance of NEAT1 targeting is further underlined by its synergistic effects with the most common drugs administered for MM treatment, including bortezomib, carfilzomib, and melphalan. Overall, NEAT1 silencing is associated with a chemo-sensitizing effect of both conventional and novel therapies, and its targeting could therefore represent a promising strategy for novel anti-MM therapeutic options.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48.
Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28:732–8.
Amodio N, D’Aquila P, Passarino G, Tassone P, Bellizzi D. Epigenetic modifications in multiple myeloma: recent advances on the role of DNA and histone methylation. Expert Opin Ther targets. 2017;21:91–101.
Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775–89.
Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94.
Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci 2014;51:344–57.
Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–11.
Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109.
Nobili L, Ronchetti D, Agnelli L, Taiana E, Vinci C, Neri A. Long non-coding RNAs in multiple myeloma. Genes (Basel). 2018;9:69.
Ronchetti D, Agnelli L, Pietrelli A, Todoerti K, Manzoni M, Taiana E, et al. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci Rep. 2018;8:6557.
Taiana E, Ronchetti D, Favasuli V, Todoerti K, Manzoni M, Amodio N, et al. Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica. 2019;104:e72–6.
Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell Proliferation. 2017;50:e12329.
Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018;32:1948–57.
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26.
Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–9.
Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–8.
Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–61.
Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–34.
Roux BT, Lindsay MA, Heward JA. Knockdown of nuclear-located enhancer RNAs and long ncRNAs using locked nucleic acid gapmeRs. Methods Mol Biol. 2017;1468:11–18.
Morelli E, Biamonte L, Federico C, Amodio N, Di Martino MT, Gallo Cantafio ME, et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-miR-17-92. Blood. 2018;132:1050–63.
Nickoloff JA, Jones D, Lee SH, Williamson EA, Hromas R. Drugging the cancers addicted to DNA repair. J Natl Cancer Inst. 2017;109:djx059.
Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–43.
Xu H, Li J, Zhou ZG. NEAT1 promotes cell proliferation in multiple myeloma by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:6403–11.
Herrero AB, Gutierrez NC. Targeting ongoing DNA damage in multiple myeloma: effects of DNA damage response inhibitors on plasma cell survival. Front Oncol. 2017;7:98.
McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15.
Pawlyn C, Loehr A, Ashby C, Tytarenko R, Deshpande S, Sun J, et al. Loss of heterozygosity as a marker of homologous repair deficiency in multiple myeloma: a role for PARP inhibition? Leukemia. 2018;32:1561–6.
Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol cell. 2014;25:169–83.
Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007;67:7395–405.
Acknowledgements
This work was financially supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC) to Antonino Neri (IG16722, IG10136, and the “Special Program Molecular Clinical Oncology-5 per mille” no. 9980, 2010/15); Elisa Taiana was supported by a fellowship (#19370) from Fondazione Italiana Ricerca sul cancro (FIRC); Katia Todoerti was supported by a fellowship from Fondazione Umberto Veronesi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Taiana, E., Favasuli, V., Ronchetti, D. et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 34, 234–244 (2020). https://doi.org/10.1038/s41375-019-0542-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0542-5
This article is cited by
-
Targeting and engineering long non-coding RNAs for cancer therapy
Nature Reviews Genetics (2024)
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies
Signal Transduction and Targeted Therapy (2023)
-
Knockdown of SIRLNT by antisense LNA GapmeRs and evaluation of its inhibitory effect on proliferation and apoptosis in MCF7 cell line
Indian Journal of Gynecologic Oncology (2023)
-
Expression relationship and significance of NEAT1 and miR-27a-3p in serum and cerebrospinal fluid of patients with Alzheimer’s disease
BMC Neurology (2022)
-
CRISPR/Cas9 gene editing: a new approach for overcoming drug resistance in cancer
Cellular & Molecular Biology Letters (2022)