Abstract
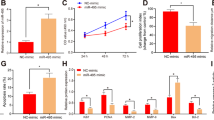
The exact mechanism of preeclampsia (PE) remains unclear, accumulating researches have indicated multiple epigenetic factors relate to PE and histone methylation plays a crucial role in modifying the gene expression. So we aimed to confirm that abnormal expression of histone demethylase JHDM1D contributes to PE and lower expression of HLA-G in PE. We tested the expression of JHDM1D, H3K9me2, and H3K27me2 in the placentas of PE and normal control (NC)women who had a healthy pregnancy with Immunohistochemistry and we found that JHDM1D, H3K9me2, and H3K27me2 were all mainly expressed in the nuclei of the extra-villous trophoblasts (EVTs). JHDM1D was lower expressed in PE than in NC placentas, corresponding with the mRNA level and protein level with qTR-PCR and western blot, while H3K9me2 and H3K27me2 were higher expressed in PE. We further investigated the biological functions of JHDM1D in HTR-8/SVneo cells. We found that siJHDM1D inhibited cell growth after 24 h of the transfection and reduced the invasion, while increasing the apoptosis of HTR-8/SVneo. We then constructed the siJHDM1D stable cell line and confirmed with CHIP-qPCR that siJHDM1D inhibited the expression of HLA-G through increased the enrichment of H3K9me2 and H3K27me2 in the JHDM1D bounding region of HLA-G. Taken together, our study confirms that decreased expression of JHDM1D is associated with PE through down-regulating HLA-G and casts new light to the diagnosis and therapy of PE.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 2009;33:130–7.
Liu X, Gu W, Li X. HLA-G regulates the invasive properties of JEG-3 choriocarcinoma cells by controlling STAT3 activation. Placenta. 2013;34:1044–52.
Chen LJ, Han ZQ, Zhou H, Zou L, Zou P. Inhibition of HLA-G expression via RNAi abolishes resistance of extravillous trophoblast cell line TEV-1 to NK lysis. Placenta. 2010;31:519–27.
Liu H, Liu X, Jin H, Yang F, Gu W, Li X. Proteomic analysis of knock-down HLA-G in invasion of human trophoblast cell line JEG-3. Int J Clin Exp Pathol. 2013;6:2451–9.
Yie SM, Li LH, Li YM, Librach C. HLA-G protein concentrations in maternal serum and placental tissue are decreased in preeclampsia. Am J Obstet Gynecol. 2004;191:525–9.
Hara N, Fujii T, Yamashita T, Kozuma S, Okai T, Taketani Y. Altered expression of human leukocyte antigen G (HLA-G) on extravillous trophoblasts in preeclampsia: immunohistological demonstration with anti-HLA-G specific antibody “87G” and anti-cytokeratin antibody “CAM5.2”. Am J Reprod Immunol. 1996;36:349–58.
Tang Y, Liu H, Li H, Peng T, Gu W, Li X. Hypermethylation of the HLA-G promoter is associated with preeclampsia. Mol Hum Reprod. 2015;21:736–44.
Jasinski-Bergner S, Reches A, Stoehr C, Massa C, Gonschorek E, Huettelmaier S, et al. Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget. 2016;7:26866–78.
Holling TM, Bergevoet MW, Wierda RJ, van Eggermond MC, van den Elsen PJ. Genetic and epigenetic control of the major histocompatibility complex class Ib gene HLA-G in trophoblast cell lines. Ann N Y Acad Sci. 2009;1173:538–44.
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6.
Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–7.
Costa-Reis P, Sullivan KE. Genetics and epigenetics of systemic lupus erythematosus. Curr Rheumatol Rep. 2013;15:369.
ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. American College of Obstetricians and Gynecologists. Int J Gynaecol Obstet. 2002;77:67–75.
Song X, Rui C, Meng L, Zhang R, Shen R, Ding H, et al. Long non-coding RNA RPAIN regulates the invasion and apoptosis of trophoblast cell lines via complement protein C1q. Oncotarget. 2017;8:7637–46.
Qian Y, Lu Y, Rui C, Qian Y, Cai M, Jia R. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol Biochem. 2016;39:1380–90.
Zhuang XW, Li J, Brost BC, Xia XY, Chen HB, Wang CX, et al. Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia. Curr Pharm Des. 2014;20:1796–802.
Luo S, Cao N, Tang Y, Gu W. Identification of key microRNAs and genes in preeclampsia by bioinformatics analysis. PLoS One. 2017;12:e178549.
Tang Y, Liu H, Li H, Peng T, Gu W, Li X. Hypermethylation of the HLA-G promoter is associated with preeclampsia. Mol Hum Reprod. 2015;21:736–44.
Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12:1065–74.
Osawa T, Muramatsu M, Wang F, Tsuchida R, Kodama T, Minami T, et al. Increased expression of histone demethylase JHDM1D under nutrient starvation suppresses tumor growth via down-regulating angiogenesis. Proc Natl Acad Sci USA. 2011;108:20725–9.
Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–6.
Ji X, Jin S, Qu X, Li K, Wang H, He H, et al. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC Cancer. 2015;15:801.
Xia M, Yao L, Zhang Q, Wang F, Mei H, Guo X, et al. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget. 2017;8:19795–802.
Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–7.
Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FM, Timmers HT, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30:3286–98.
Johansson C, Tumber A, Che K, Cain P, Nowak R, Gileadi C, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics. 2014;6:89–120.
Lin H, Wang Y, Wang Y, Tian F, Pu P, Yu Y, et al. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 2010;20:899–907.
Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43.
Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26:1364–75.
Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–7.
Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2:1849–61.
Funding
This work was supported by National Key R&D Program of China (2016YFC1000403) and by Natural Science Foundation of China (Grant No. 81471470 to W.-R.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Luo, S., Pei, J., Li, X. et al. Decreased expression of JHDMID in placenta is associated with preeclampsia through HLA-G. J Hum Hypertens 32, 448–454 (2018). https://doi.org/10.1038/s41371-018-0062-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41371-018-0062-1